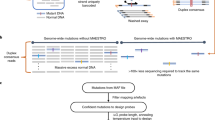
Abstract
A new generation of technologies is poised to reduce DNA sequencing costs by several orders of magnitude. But our ability to fully leverage the power of these technologies is crippled by the absence of suitable 'front-end' methods for isolating complex subsets of a mammalian genome at a scale that matches the throughput at which these platforms will routinely operate. We show that targeting oligonucleotides released from programmable microarrays can be used to capture and amplify ∼10,000 human exons in a single multiplex reaction. Additionally, we show integration of this protocol with ultra-high-throughput sequencing for targeted variation discovery. Although the multiplex capture reaction is highly specific, we found that nonuniform capture is a key issue that will need to be resolved by additional optimization. We anticipate that highly multiplexed methods for targeted amplification will enable the comprehensive resequencing of human exons at a fraction of the cost of whole-genome resequencing.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
21 October 2007
In the version of this article initially published online,the affiliation for Jay Shendure was listed as Department of Computer Science, Virginia Commonwealth University,601 West Main Street,Richmond,Virginia 23284,USA. The correct affiliation should be Department of Genome Sciences,University of Washington,1705 NE Pacific St.,Seattle,Washington 98195,USA. The error has been corrected for all versions of the article.
References
Shendure, J., Mitra, R.D., Varma, C. & Church, G.M. Advanced sequencing technologies: methods and goals. Nat. Rev. Genet. 5, 335–344 (2004).
Shendure, J. et al. Accurate multiplex polony sequencing of an evolved bacterial genome. Science 309, 1728–1732 (2005).
Johnson, D.S., Mortazavi, A., Myers, R.M. & Wold, B. Genome-wide mapping of in vivo protein-DNA interactions. Science 316, 1497–1502 (2007).
Margulies, M. et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 376–380 (2005).
Stephens, M., Sloan, J.S., Robertson, P.D., Scheet, P. & Nickerson, D.A. Automating sequence-based detection and genotyping of SNPs from diploid samples. Nat. Genet. 38, 375–381 (2006).
Sjoblom, T. et al. The consensus coding sequences of human breast and colorectal cancers. Science 314, 268–274 (2006).
Edwards, M.C. & Gibbs, R.A. Multiplex PCR: advantages, development, and applications. PCR Methods Appl. 3, S65–S75 (1994).
Markoulatos, P., Siafakas, N. & Moncany, M. Multiplex polymerase chain reaction: a practical approach. J. Clin. Lab. Anal. 16, 47–51 (2002).
Hardenbol, P. et al. Multiplexed genotyping with sequence-tagged molecular inversion probes. Nat. Biotechnol. 21, 673–678 (2003).
Hardenbol, P. et al. Highly multiplexed molecular inversion probe genotyping: over 10,000 targeted SNPs genotyped in a single tube assay. Genome Res. 15, 269–275 (2005).
Dahl, F. et al. Multigene amplification and massively parallel sequencing for cancer mutation discovery. Proc. Natl. Acad. Sci. USA 104, 9387–9392 (2007).
Fredriksson, S. et al. Multiplex amplification of all coding sequences within 10 cancer genes by Gene-Collector. Nucleic Acids Res. 35, e47 (2007).
Bashiardes, S. et al. Direct genomic selection. Nat. Methods 2, 63–69 (2005).
Okou, D.T. et al. Microarray-based genomic selection for high-throughput resequencing. Nat. Methods, advance online publication 14 October 2007 (doi:10.1038/nmeth1109).
Albert, T.J. et al. Direct selection of human genomic loci by microarray hybridization. Nat. Methods, advance online publication 14 October 2007 (doi:10.1038/nmeth1111).
Tian, J. et al. Accurate multiplex gene synthesis from programmable DNA microchips. Nature 432, 1050–1054 (2004).
Dahl, F., Gullberg, M., Stenberg, J., Landegren, U. & Nilsson, M. Multiplex amplification enabled by selective circularization of large sets of genomic DNA fragments. Nucleic Acids Res. 33, e71 (2005).
Cohen, J. et al. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat. Genet. 37, 161–165 (2005).
Farooqi, I.S. et al. Clinical and molecular genetic spectrum of congenital deficiency of the leptin receptor. N. Engl. J. Med. 356, 237–247 (2007).
Romeo, S. et al. Population-based resequencing of ANGPTL4 uncovers variations that reduce triglycerides and increase HDL. Nat. Genet. 39, 513–516 (2007).
Topol, E.J. & Frazer, K.A. The resequencing imperative. Nat. Genet. 39, 439–440 (2007).
Futreal, P.A. et al. A census of human cancer genes. Nat. Rev. Cancer 4, 177–183 (2004).
Greenman, C. et al. Patterns of somatic mutation in human cancer genomes. Nature 446, 153–158 (2007).
Collins, F.S. & Barker, A.D. Mapping the cancer genome. Pinpointing the genes involved in cancer will help chart anew course across the complex landscape of human malignancies. Sci. Am. 296, 50–57 (2007).
Acknowledgements
This work was supported by a Center for Excellence in Genome Sciences grant from the National Human Genome Research Institute, and a SPARC grant from the Broad Institute of Massachusetts Institute of Technology and Harvard University. We are grateful to G. Buck, M. Davis, N. Sheth, C. Childress, Jr. and J. Noble (Center for High Performance Computing and Center for the Study of Biological Complexity, Virginia Commonwealth University) for setting up the Illumina Genome Analyzer analysis pipeline. We thank H. Ji, S. Fredriksson, A. Gnirke, E. Lander, D. Jaffe and C. Nusbaum for discussions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
E.M.L. and B.J.P. are employed by Agilent Technologies, Inc., and Agilent reagents are used in the research presented in this article.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3, Supplementary Table 1, Supplementary Methods (PDF 271 kb)
Supplementary Data 1
Sequences of targeting oligonucleotides and targets (55,000-plex). (TXT 12318 kb)
Supplementary Data 2
Sequences of targeting oligonucleotides and targets (480-plex). (TXT 93 kb)
Rights and permissions
About this article
Cite this article
Porreca, G., Zhang, K., Li, J. et al. Multiplex amplification of large sets of human exons. Nat Methods 4, 931–936 (2007). https://doi.org/10.1038/nmeth1110
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth1110
This article is cited by
-
Allele mining through TILLING and EcoTILLING approaches in vegetable crops
Planta (2023)
-
DNA methylation markers in the diagnosis and prognosis of common leukemias
Signal Transduction and Targeted Therapy (2020)
-
A novel emulsion PCR method coupled with fluorescence spectrophotometry for simultaneous qualitative, quantitative and high-throughput detection of multiple DNA targets
Scientific Reports (2019)
-
Intergenic disease-associated regions are abundant in novel transcripts
Genome Biology (2017)
-
Genomic innovation for crop improvement
Nature (2017)