Abstract
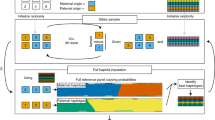
We developed a general method, microarray-based genomic selection (MGS), capable of selecting and enriching targeted sequences from complex eukaryotic genomes without the repeat blocking steps necessary for bacterial artificial chromosome (BAC)-based genomic selection. We demonstrate that large human genomic regions, on the order of hundreds of kilobases, can be enriched and resequenced with resequencing arrays. MGS, when combined with a next-generation resequencing technology, can enable large-scale resequencing in single-investigator laboratories.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Cutler, D.J. et al. Genome Res. 11, 1913–1925 (2001).
Margulies, M. et al. Nature 437, 376–380 (2005).
Shendure, J., Mitra, R.D., Varma, C. & Church, G.M. Nat. Rev. Genet. 5, 335–344 (2004).
Shendure, J. et al. Science 309, 1728–1732 (2005).
Zwick, M.E. et al. Genome Biol. 6, R10 (2005).
Hinds, D.A. et al. Science 307, 1072–1079 (2005).
Sjoblom, T. et al. Science 314, 268–274 (2006).
Raymond, C.K. et al. Genomics 86, 759–766 (2005).
Raymond, C.K., Sims, E.H. & Olson, M.V. Genome Res. 12, 190–197 (2002).
Kouprina, N., Noskov, V.N. & Larionov, V. Methods Mol. Biol. 349, 85–101 (2006).
Dahl, F. et al. Proc. Natl. Acad. Sci. USA 104, 9387–9392 (2007).
Bashiardes, S. et al. Nat. Methods 2, 63–69 (2005).
De Boulle, K. et al. Nat. Genet. 3, 31–35 (1993).
Gu, Y., Lugenbeel, K.A., Vockley, J.G., Grody, W.W. & Nelson, D.L. Hum. Mol. Genet. 3, 1705–1706 (1994).
Porreca, G.J. et al. Nat. Methods, advance online publication 14 October 2007 (doi:10.1038/nmeth1110).
Albert, T.J. et al. Nat. Methods, advance online publication 14 October 2007 (doi:10.1038/nmeth1111).
Bentley, D.R. Curr. Opin. Genet. Dev. 16, 545–552 (2006).
Acknowledgements
Funding for this work was provided by the US National Institutes of Health (NIH) National Institute of Mental Health RO1 MH076439-03 (MEZ) and by the Gift Fund, NIH.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
T.J.A. and C.M. are employees of NimbleGen Systems, Inc.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–2, Supplementary Methods, Supplementary Table 1 (PDF 1776 kb)
Supplementary Data 1
Oligonucleotide Capture Probes for 50kb FMR1 Genomic Region (DOC 25359 kb)
Supplementary Data 2
Oligonucleotide Capture Probes for 304kb FMR1/FMR2 Genomic Region (DOC 28891 kb)
Supplementary Data 3
DNA Sequence Data for 50kb Region (ZIP 65 kb)
Supplementary Data 4
DNA Sequence Data for 304kb Region (ZIP 1893 kb)
Rights and permissions
About this article
Cite this article
Okou, D., Steinberg, K., Middle, C. et al. Microarray-based genomic selection for high-throughput resequencing. Nat Methods 4, 907–909 (2007). https://doi.org/10.1038/nmeth1109
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth1109
This article is cited by
-
EYS is a major gene involved in retinitis pigmentosa in Japan: genetic landscapes revealed by stepwise genetic screening
Scientific Reports (2020)
-
Gene Saturation: An Approach to Assess Exploration Stage of Gene Interaction Networks
Scientific Reports (2019)
-
High-sensitivity HLA typing by Saturated Tiling Capture Sequencing (STC-Seq)
BMC Genomics (2018)
-
Methylation-based enrichment facilitates low-cost, noninvasive genomic scale sequencing of populations from feces
Scientific Reports (2018)