Abstract
We have assembled de novo the Escherichia coli K-12 MG1655 chromosome in a single 4.6-Mb contig using only nanopore data. Our method has three stages: (i) overlaps are detected between reads and then corrected by a multiple-alignment process; (ii) corrected reads are assembled using the Celera Assembler; and (iii) the assembly is polished using a probabilistic model of the signal-level data. The assembly reconstructs gene order and has 99.5% nucleotide identity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Jain, M. et al. Nat. Methods 12, 351–356 (2015).
Koren, S. et al. Genome Biol. 14, R101 (2013).
Koren, S. et al. Nat. Biotechnol. 30, 693–700 (2012).
Rasko, D.A. et al. N. Engl. J. Med. 365, 709–717 (2011).
Chin, C.-S. et al. Nat. Methods 10, 563–569 (2013).
Kim, K.E. et al. Sci. Data 1, 140045 (2014).
Koren, S. & Phillippy, A.M. Curr. Opin. Microbiol. 23, 110–120 (2015).
Quick, J., Quinlan, A.R. & Loman, N.J. Gigascience 3, 22 (2014).
Goodwin, S. et al. Preprint at bioRxiv 10.1101/013490 (2015).
Loman, N.J. & Quinlan, A.R. Bioinformatics 30, 3399–3401 (2014).
Myers, G. in Int. Workshop Algorithms Bioinformatics (eds. Brown, D. & Morgenstern, B.) 52–67 (Springer, 2014).
Lee, C., Grasso, C. & Sharlow, M.F. Bioinformatics 18, 452–464 (2002).
Myers, E.W. et al. Science 287, 2196–2204 (2000).
Gurevich, A., Saveliev, V., Vyahhi, N. & Tesler, G. Bioinformatics 29, 1072–1075 (2013).
Darling, A.E., Mau, B. & Perna, N.T. PLoS ONE 5, e11147 (2010).
Milne, I. et al. Brief. Bioinform. 14, 193–202 (2013).
Treangen, T.J., Sommer, D.D., Angly, F.E., Koren, S. & Pop, M. Curr. Protoc. Bioinformatics 33, 11.8 (2011).
Delcher, A.L., Phillippy, A., Carlton, J. & Salzberg, S.L. Nucleic Acids Res. 30, 2478–2483 (2002).
Li, H. Preprint at http://arxiv.org/abs/1303.3997 (2013).
Quinlan, A.R. & Hall, I.M. Bioinformatics 26, 841–842 (2010).
Cock, P.J.A. et al. Bioinformatics 25, 1422–1423 (2009).
Acknowledgements
Data analysis was performed on the Medical Research Council Cloud Infrastructure for Microbial Bioinformatics (CLIMB) cyberinfrastructure. N.J.L. is funded by a Medical Research Council Special Training Fellowship in Biomedical Informatics. J.Q. is funded by the UK National Institute for Health Research (NIHR) Surgical Reconstruction and Microbiology Research Centre. J.T.S. is supported by the Ontario Institute for Cancer Research through funding provided by the Government of Ontario. We thank the staff of Oxford Nanopore for technical help and advice during the MinION Access Programme. We are grateful to the EU COST action ES1103, whose funding allowed us to attend a hackathon that kick-started the work presented here. We thank L. Parts for comments on the manuscript and H. Eno for help with proofreading.
Author information
Authors and Affiliations
Contributions
N.J.L. and J.T.S. conceived the project. N.J.L., J.Q. and J.T.S. implemented the Nanocorrect pipeline. J.T.S. conceived and implemented the Nanopolish pipeline. J.Q. generated the nanopore E. coli sequence data. N.J.L. and J.T.S. performed de novo assembly and analyzed the results. N.J.L. and J.T.S. wrote the manuscript. All authors approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
N.J.L. and J.T.S. are members of the MinION Access Programme (MAP). N.J.L. has received free-of-charge reagents for nanopore sequencing presented in this study. N.J.L., J.Q. and J.T.S. have received travel and accommodation expenses to speak at an Oxford Nanopore–organized symposium. N.J.L. and J.Q. have ongoing research collaborations with Oxford Nanopore but do not receive financial compensation for this.
Integrated supplementary information
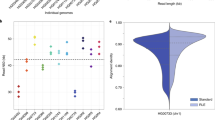
Supplementary Figure 1 Kernel density plot showing the accuracy of reads from the four individual MinION runs used to generate the de novo assembly.
The mean accuracy varies from 78.2% (run 3) to 82.2% (run 1).
Supplementary Figure 2 Kernel density plot demonstrating the raw nanopore read accuracy and effect of two rounds of error correction on accuracy.
The mauve area represents uncorrected sequencing reads, where the green area shows the improvement in accuracy after the first round of correction and the yellow shows improvement from the second round of correction. Further rounds of correction did not improve the accuracy further.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3, Supplementary Tables 1 and 2 and Supplementary Note (PDF 785 kb)
Rights and permissions
About this article
Cite this article
Loman, N., Quick, J. & Simpson, J. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat Methods 12, 733–735 (2015). https://doi.org/10.1038/nmeth.3444
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.3444
This article is cited by
-
Re-analysis of an outbreak of Shiga toxin-producing Escherichia coli O157:H7 associated with raw drinking milk using Nanopore sequencing
Scientific Reports (2024)
-
N6-methyladenosine-mediated feedback regulation of abscisic acid perception via phase-separated ECT8 condensates in Arabidopsis
Nature Plants (2024)
-
Flexible and efficient handling of nanopore sequencing signal data with slow5tools
Genome Biology (2023)
-
A targeted long-read sequencing approach questions the association of OXTR methylation with high-functioning autism
Clinical Epigenetics (2023)
-
Fine-scale genomic tracking of Ross River virus using nanopore sequencing
Parasites & Vectors (2023)