Abstract
NOTE: In the version of this Brief Communication initially published, an author name (Lukas Mueller) was incorrect. The correct author name is Lukas N Mueller. The error has been corrected in the HTML and PDF versions of the article.
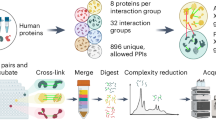
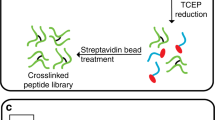
We describe a method to identify cross-linked peptides from complex samples and large protein sequence databases by combining isotopically tagged cross-linkers, chromatographic enrichment, targeted proteomics and a new search engine called xQuest. This software reduces the search space by an upstream candidate-peptide search before the recombination step. We showed that xQuest can identify cross-linked peptides from a total Escherichia coli lysate with an unrestricted database search.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Change history
27 June 2008
In the version of this Brief Communication initially published, an author name (Lukas Mueller) was incorrect. The correct author name is Lukas N Mueller. The error has been corrected in the HTML and PDF versions of the article.
References
Young, M.M. et al. Proc. Natl. Acad. Sci. USA 97, 5802–5806 (2000).
Sinz, A. J. Mass Spectrom. 38, 1225–1237 (2003).
Seebacher, J. et al. J. Proteome Res. 5, 2270–2282 (2006).
Ihling, C. et al. J. Am. Soc. Mass Spectrom. 17, 1100–1113 (2006).
Muller, D.R. et al. Anal. Chem. 73, 1927–1934 (2001).
Dihazi, G.H. & Sinz, A. Rapid Commun. Mass Spectrom. 17, 2005–2014 (2003).
Huang, B.X., Kim, H.Y. & Dass, C. J. Am. Soc. Mass Spectrom. 15, 1237–1247 (2004).
Novak, P. et al. Anal. Chem. 77, 5101–5106 (2005).
Pearson, K.M., Pannell, L.K. & Fales, H.M. Rapid Commun. Mass Spectrom. 16, 149–159 (2002).
Gao, Q. et al. Anal. Chem. 78, 2145–2149 (2006).
Schilling, B., Row, R.H., Gibson, B.W., Guo, X. & Young, M.M. J. Am. Soc. Mass Spectrom. 14, 834–850 (2003).
de Koning, L.J. et al. FEBS J. 273, 281–291 (2006).
Back, J.W., de Jong, L., Muijsers, A.O. & de Koster, C.G. J. Mol. Biol. 331, 303–313 (2003).
Sinz, A., Kalkhof, S. & Ihling, C. J. Am. Soc. Mass Spectrom. 16, 1921–1931 (2005).
Trester-Zedlitz, M. et al. J. Am. Chem. Soc. 125, 2416–2425 (2003).
Acknowledgements
O.R. was supported by fellowships of the Deutsche Forschungsgemeinschaft and the Roche Research Foundation; M.B. was supported by a long-term fellowship of the European Molecular Biology Organization and a Marie Curie fellowship of the European Commission. A.S. was supported in part by a grant from F. Hoffmann–La Roche Ltd provided to the Competence Center for Systems Physiology and Metabolic Disease. The work was supported in part with Federal funds from the US National Heart, Lung and Blood Institute, National Institutes of Health (contract N01-HV-28179) and with funds from ETH Zurich.
Author information
Authors and Affiliations
Contributions
O.R. was responsible for most of the software design and programming, conducted most of the experiments and most of the data evaluation; J.S. synthesized the cross-linker and contributed to software design; T.W. contributed to scoring, data evaluation and part of experiments; L.N.M. analyzed and aligned MS1 data; M.B. performed part of experiments and evaluated data; A.S. designed mass spectrometric methods for analyzing cross-linked samples; M.M. contributed to scoring; R.A. coordinated the project, contributed ideas and concept.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5, Supplementary Tables 1–2, Supplementary Results, Supplementary Discussion, Supplementary Methods (PDF 1982 kb)
Rights and permissions
About this article
Cite this article
Rinner, O., Seebacher, J., Walzthoeni, T. et al. Identification of cross-linked peptides from large sequence databases. Nat Methods 5, 315–318 (2008). https://doi.org/10.1038/nmeth.1192
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1192
This article is cited by
-
FTD-tau S320F mutation stabilizes local structure and allosterically promotes amyloid motif-dependent aggregation
Nature Communications (2023)
-
Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin
Nature Communications (2023)
-
Targeted cross-linker delivery for the in situ mapping of protein conformations and interactions in mitochondria
Nature Communications (2023)
-
Biophysical properties of a tau seed
Scientific Reports (2021)
-
DnaJC7 binds natively folded structural elements in tau to inhibit amyloid formation
Nature Communications (2021)