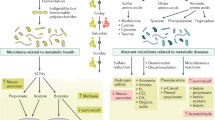
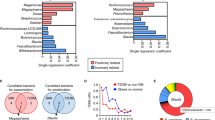
Abstract
Metformin is widely used in the treatment of type 2 diabetes (T2D), but its mechanism of action is poorly defined. Recent evidence implicates the gut microbiota as a site of metformin action. In a double-blind study, we randomized individuals with treatment-naive T2D to placebo or metformin for 4 months and showed that metformin had strong effects on the gut microbiome. These results were verified in a subset of the placebo group that switched to metformin 6 months after the start of the trial. Transfer of fecal samples (obtained before and 4 months after treatment) from metformin-treated donors to germ-free mice showed that glucose tolerance was improved in mice that received metformin-altered microbiota. By directly investigating metformin–microbiota interactions in a gut simulator, we showed that metformin affected pathways with common biological functions in species from two different phyla, and many of the metformin-regulated genes in these species encoded metalloproteins or metal transporters. Our findings provide support for the notion that altered gut microbiota mediates some of metformin's antidiabetic effects.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Nathan, D.M. et al. Medical management of hyperglycemia in type 2 diabetes: a consensus algorithm for the initiation and adjustment of therapy: a consensus statement of the American Diabetes Association and the European Association for the Study of Diabetes. Diabetes Care 32, 193–203 (2009).
Pernicova, I. & Korbonits, M. Metformin—mode of action and clinical implications for diabetes and cancer. Nat. Rev. Endocrinol. 10, 143–156 (2014).
Zhou, G. et al. Role of AMP-activated protein kinase in mechanism of metformin action. J. Clin. Invest. 108, 1167–1174 (2001).
Shaw, R.J. et al. The kinase LKB1 mediates glucose homeostasis in liver and therapeutic effects of metformin. Science 310, 1642–1646 (2005).
Fullerton, M.D. et al. Single phosphorylation sites in Acc1 and Acc2 regulate lipid homeostasis and the insulin-sensitizing effects of metformin. Nat. Med. 19, 1649–1654 (2013).
Foretz, M. et al. Metformin inhibits hepatic gluconeogenesis in mice independently of the LKB1/AMPK pathway via a decrease in hepatic energy state. J. Clin. Invest. 120, 2355–2369 (2010).
Madiraju, A.K. et al. Metformin suppresses gluconeogenesis by inhibiting mitochondrial glycerophosphate dehydrogenase. Nature 510, 542–546 (2014).
Miller, R.A. et al. Biguanides suppress hepatic glucagon signalling by decreasing production of cyclic AMP. Nature 494, 256–260 (2013).
McCreight, L.J., Bailey, C.J. & Pearson, E.R. Metformin and the gastrointestinal tract. Diabetologia 59, 426–435 (2016).
Duca, F.A. et al. Metformin activates a duodenal Ampk-dependent pathway to lower hepatic glucose production in rats. Nat. Med. 21, 506–511 (2015).
Stepensky, D., Friedman, M., Raz, I. & Hoffman, A. Pharmacokinetic-pharmacodynamic analysis of the glucose-lowering effect of metformin in diabetic rats reveals first-pass pharmacodynamic effect. Drug Metab. Dispos. 30, 861–868 (2002).
Buse, J.B. et al. The primary glucose-lowering effect of metformin resides in the gut, not the circulation. Results from short-term pharmacokinetic and 12-week dose-ranging studies. Diabetes Care 39, 198–205 (2016).
Shin, N.R. et al. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut 63, 727–735 (2014).
Zhang, X. et al. Modulation of gut microbiota by berberine and metformin during the treatment of high-fat diet-induced obesity in rats. Sci. Rep. 5, 14405 (2015).
Lee, H. & Ko, G. Effect of metformin on metabolic improvement and gut microbiota. Appl. Environ. Microbiol. 80, 5935–5943 (2014).
Karlsson, F.H. et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 498, 99–103 (2013).
Forslund, K. et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 528, 262–266 (2015).
de la Cuesta-Zuluaga, J. et al. Metformin is associated with higher relative abundance of mucin-degrading Akkermansia muciniphila and several short-chain fatty acid–producing microbiota in the gut. Diabetes Care 40, 54–62 (2017).
Karlsson, F.H., Nookaew, I. & Nielsen, J. Metagenomic data utilization and analysis (MEDUSA) and construction of a global gut microbial gene catalogue. PLoS Comput. Biol. 10, e1003706 (2014).
Everard, A. et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA 110, 9066–9071 (2013).
Plovier, H. et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat. Med. 23, 107–113 (2017).
Dao, M.C. et al. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: relationship with gut microbiome richness and ecology. Gut 65, 426–436 (2016).
Park, S.K., Kim, M.S., Roh, S.W. & Bae, J.W. Blautia stercoris sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 62, 776–779 (2012).
Korem, T. et al. Growth dynamics of gut microbiota in health and disease inferred from single metagenomic samples. Science 349, 1101–1106 (2015).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Wong, J.M., de Souza, R., Kendall, C.W., Emam, A. & Jenkins, D.J. Colonic health: fermentation and short chain fatty acids. J. Clin. Gastroenterol. 40, 235–243 (2006).
Koh, A., De Vadder, F., Kovatcheva-Datchary, P. & Bäckhed, F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 165, 1332–1345 (2016).
Ridlon, J.M., Harris, S.C., Bhowmik, S., Kang, D.J. & Hylemon, P.B. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes 7, 22–39 (2016).
Caspary, W.F. et al. Alteration of bile acid metabolism and vitamin-B12-absorption in diabetics on biguanides. Diabetologia 13, 187–193 (1977).
Scarpello, J.H., Hodgson, E. & Howlett, H.C. Effect of metformin on bile salt circulation and intestinal motility in type 2 diabetes mellitus. Diabet. Med. 15, 651–656 (1998).
Zhernakova, A. et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 352, 565–569 (2016).
Cotillard, A. et al. Dietary intervention impact on gut microbial gene richness. Nature 500, 585–588 (2013).
Chen, J., Wang, R., Li, X.F. & Wang, R.L. Bifidobacterium adolescentis supplementation ameliorates visceral fat accumulation and insulin sensitivity in an experimental model of the metabolic syndrome. Br. J. Nutr. 107, 1429–1434 (2012).
Desai, M.S. et al. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell 167, 1339–1353.e21 (2016).
Roopchand, D.E. et al. Dietary polyphenols promote growth of the gut bacterium Akkermansia muciniphila and attenuate high-fat diet-induced metabolic syndrome. Diabetes 64, 2847–2858 (2015).
Anhê, F.F. et al. A polyphenol-rich cranberry extract protects from diet-induced obesity, insulin resistance and intestinal inflammation in association with increased Akkermansia spp. population in the gut microbiota of mice. Gut 64, 872–883 (2015).
Greer, R.L. et al. Akkermansia muciniphila mediates negative effects of IFNγ on glucose metabolism. Nat. Commun. 7, 13329 (2016).
Zhang, H., Sparks, J.B., Karyala, S.V., Settlage, R. & Luo, X.M. Host adaptive immunity alters gut microbiota. ISME J. 9, 770–781 (2015).
Collado, M.C., Derrien, M., Isolauri, E., de Vos, W.M. & Salminen, S. Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl. Environ. Microbiol. 73, 7767–7770 (2007).
Kong, F. et al. Gut microbiota signatures of longevity. Curr. Biol. 26, R832–R833 (2016).
Tremaroli, V. et al. Roux-en-Y gastric bypass and vertical banded gastroplasty induce long-term changes on the human gut microbiome contributing to fat mass regulation. Cell Metab. 22, 228–238 (2015).
Everard, A. et al. Microbiome of prebiotic-treated mice reveals novel targets involved in host response during obesity. ISME J. 8, 2116–2130 (2014).
Fernández-Real, J.M. & Manco, M. Effects of iron overload on chronic metabolic diseases. Lancet Diabetes Endocrinol. 2, 513–526 (2014).
Logie, L. et al. Cellular responses to the metal-binding properties of metformin. Diabetes 61, 1423–1433 (2012).
Wahlström, A. et al. Induction of farnesoid X receptor signaling in germ-free mice colonized with a human microbiota. J. Lipid Res. 58, 412–419 (2017).
Wahlström, A., Sayin, S.I., Marschall, H.U. & Bäckhed, F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. 24, 41–50 (2016).
Schaap, F.G., Trauner, M. & Jansen, P.L. Bile acid receptors as targets for drug development. Nat. Rev. Gastroenterol. Hepatol. 11, 55–67 (2014).
De Vadder, F. et al. Microbiota-produced succinate improves glucose homeostasis via intestinal gluconeogenesis. Cell Metab. 24, 151–157 (2016).
Bäckhed, F., Manchester, J.K., Semenkovich, C.F. & Gordon, J.I. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc. Natl. Acad. Sci. USA 104, 979–984 (2007).
Vioque, J. et al. Reproducibility and validity of a food frequency questionnaire among pregnant women in a Mediterranean area. Nutr. J. 12, 26 (2013).
American Diabetes Association. Diagnosis and classification of diabetes mellitus. Diabetes Care 33 (Suppl. 1), S62–S69 (2010).
Langmead, B. & Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Love, M.I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Young, M.D., Wakefield, M.J., Smyth, G.K. & Oshlack, A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11, R14 (2010).
Kindt, R. & Coe, R. Tree diversity analysis. A manual and software for common statistical methods for ecological and biodiversity studies (World Agroforestry Centre, Nairobi, Kenya, 2005).
McMurdie, P.J. & Holmes, S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8, e61217 (2013).
Csárdi, G. & Nepusz, T. The igraph software package for complex network research. InterJournal. Complex Syst. 1695, 1695 (2006).
Altschul, S.F., Gish, W., Miller, W., Myers, E.W. & Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Tatusova, T., Ciufo, S., Fedorov, B., O'Neill, K. & Tolstoy, I. RefSeq microbial genomes database: new representation and annotation strategy. Nucleic Acids Res. 42, D553–D559 (2014).
Li, J. et al. An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 32, 834–841 (2014).
Wichmann, A. et al. Microbial modulation of energy availability in the colon regulates intestinal transit. Cell Host Microbe 14, 582–590 (2013).
Lee, Y.S. et al. Insulin-like peptide 5 is a microbially regulated peptide that promotes hepatic glucose production. Mol. Metab. 5, 263–270 (2016).
Van den Abbeele, P. et al. Microbial community development in a dynamic gut model is reproducible, colon region specific, and selective for Bacteroidetes and Clostridium cluster IX. Appl. Environ. Microbiol. 76, 5237–5246 (2010).
Salonen, A. et al. Comparative analysis of fecal DNA extraction methods with phylogenetic microarray: effective recovery of bacterial and archaeal DNA using mechanical cell lysis. J. Microbiol. Methods 81, 127–134 (2010).
Murphy, N.R. & Hellwig, R.J. Improved nucleic acid organic extraction through use of a unique gel barrier material. Biotechniques 21, 934–936, 938–939 (1996).
Zoetendal, E.G. et al. Isolation of RNA from bacterial samples of the human gastrointestinal tract. Nat. Protoc. 1, 954–959 (2006).
Gu, Z., Gu, L., Eils, R., Schlesner, M. & Brors, B. circlize Implements and enhances circular visualization in R. Bioinformatics 30, 2811–2812 (2014).
Franceschini, A. et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41, D808–D815 (2013).
Team, R.C.R. A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/ (2015).
Champely, S. pwr: Basic Functions for Power Analysis. R package version 1.1-3. http://CRAN.R-project.org/package=pwr (2015).
Leucht, S., Helfer, B., Gartlehner, G. & Davis, J.M. How effective are common medications: a perspective based on meta-analyses of major drugs. BMC Med. 13, 253 (2015).
Oksanen, J. et al. vegan: Community Ecology Package. R package version 2.2-1 http://CRAN.R-project.org/package=vegan (2015).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995).
Acknowledgements
We thank C. Arvidsson, S. Nordin-Larsson, C. Wennberg, and U. Enqvist for superb mouse husbandry. The administrative and technical help of J.M. Moreno Navarrete, E. Huertos, M. Sabater, and O. Rovira is also acknowledged. The strain Akkermansia muciniphila DSM22959 was kindly provided by W. de Vos (Wageningen University and Helsinki University). The strain Bifidobacterium adolescentis L2-32 was kindly provided by K. Scott (The Rowett Institute of Nutrition and Health, University of Aberdeen). Whole-genome shotgun sequencing was performed at the Genomics Core Facility at the Sahlgrenska Academy, University of Gothenburg. The computations for metagenomics analyses were performed on resources provided by the Swedish National Infrastructure for Computing (SNIC) through Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX). This study was supported by the Swedish Diabetes Foundation; Swedish Research Council; Swedish Heart Lung Foundation; Torsten Söderberg's Foundation; Göran Gustafsson's Foundation; Inga Britt and Arne Lundberg's Foundation; Swedish Foundation for Strategic Research; Knut and Alice Wallenberg Foundation; the Novo Nordisk Foundation; the regional agreement on medical training and clinical research (ALF) between Region Västra Götaland and Sahlgrenska University Hospital; the Ministerio de Economía y Competitividad (PI11-00214 and PI15/01934); and FEDER funds. CIBEROBN Fisiopatología de la Obesidad y Nutrición is an initiative from the Instituto de Salud Carlos III from Spain. M.P.-F. is funded by the Obra Social Fundación la Caixa fellowship under the Severo Ochoa 2013 program. J.M.M. was supported by the Sara Borrell Fellowship from the Instituto Carlos III, EFSD/Lilly Research Fellowship and Beatriu de Pinós Fellowship from the Agency for Management of University and Research Grants (AGAUR). F.B. is a recipient of ERC Consolidator Grant (European Research Council, Consolidator grant 615362—METABASE).
Author information
Authors and Affiliations
Contributions
F.B., J.M.F.-R., V.T., and R.B. conceived and designed the study. E.E., M.P.-F., G.X., J.M.M., D.T., W.R., and J.M.F.-R. recruited cohort individuals and performed the clinical study. H.W., V.T., and F.B. conducted the bioinformatics study, analyzed all results unless otherwise indicated. H.W., V.T., R.P., and F.B. wrote the paper. M.T.K. performed the in vitro gut simulator and bacterial growth experiments. R.C. and L.M.-H. performed and analyzed the fecal microbiota transplantation experiments. M. Ståhlman performed the metabolomics experiments. M. Serino and V.T. extracted the bacterial DNA and discussed the results. L.M.O. and V.T. extracted the bacterial RNA and coordinated the metagenomics and metatranscriptomics sequencing. All authors commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4. (PDF 781 kb)
Supplementary Table 1
Diet information for the 40 subjects with T2D enrolled in this study. (XLSX 16 kb)
Supplementary Table 2
Summary of quality filtering and mapping to both the MEDUSA gene and genome catalogues. (XLSX 29 kb)
Supplementary Table 3
List of microbial strains and genera in the fecal metagenomes that were significantly increased or decreased by metformin treatment across all sampling points. (XLSX 39 kb)
Supplementary Table 4
List of KOs that were significantly affected in the fecal metagenome across all sampling points and corresponding KEGG pathway annotations. (XLSX 224 kb)
Supplementary Table 5
List of bacterial strains that were significantly increased or decreased in the gut simulator based on WGS sequencing at both DNA and RNA levels. (XLSX 15 kb)
Supplementary Table 6
List of KOs that were significantly increased or decreased in the metagenome and metatranscriptome from the in vitro gut simulator after exposure to metformin using fecal samples from donors 13 and 49 and corresponding KEGG pathway annotations. (XLSX 243 kb)
Supplementary Table 7
Annotation of the 207 and 200 genes in A. muciniphila and B. wadsworthia, respectively, that were significantly regulated by exposure to metformin in the in vitro gut simulator. (XLSX 69 kb)
Rights and permissions
About this article
Cite this article
Wu, H., Esteve, E., Tremaroli, V. et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat Med 23, 850–858 (2017). https://doi.org/10.1038/nm.4345
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.4345