Abstract
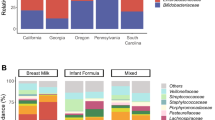
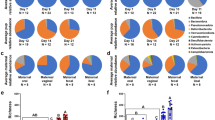
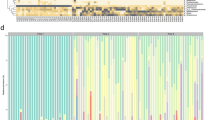
Human microbial communities are characterized by their taxonomic, metagenomic and metabolic diversity, which varies by distinct body sites and influences human physiology. However, when and how microbial communities within each body niche acquire unique taxonomical and functional signatures in early life remains underexplored. We thus sought to determine the taxonomic composition and potential metabolic function of the neonatal and early infant microbiota across multiple body sites and assess the effect of the mode of delivery and its potential confounders or modifiers. A cohort of pregnant women in their early third trimester (n = 81) were prospectively enrolled for longitudinal sampling through 6 weeks after delivery, and a second matched cross-sectional cohort (n = 81) was additionally recruited for sampling once at the time of delivery. Samples across multiple body sites, including stool, oral gingiva, nares, skin and vagina were collected for each maternal–infant dyad. Whole-genome shotgun sequencing and sequencing analysis of the gene encoding the 16S rRNA were performed to interrogate the composition and function of the neonatal and maternal microbiota. We found that the neonatal microbiota and its associated functional pathways were relatively homogeneous across all body sites at delivery, with the notable exception of the neonatal meconium. However, by 6 weeks after delivery, the infant microbiota structure and function had substantially expanded and diversified, with the body site serving as the primary determinant of the composition of the bacterial community and its functional capacity. Although minor variations in the neonatal (immediately at birth) microbiota community structure were associated with the cesarean mode of delivery in some body sites (oral gingiva, nares and skin; R2 = 0.038), this was not true for neonatal stool (meconium; Mann–Whitney P > 0.05), and there was no observable difference in community function regardless of delivery mode. For infants at 6 weeks of age, the microbiota structure and function had expanded and diversified with demonstrable body site specificity (P < 0.001, R2 = 0.189) but without discernable differences in community structure or function between infants delivered vaginally or by cesarean surgery (P = 0.057, R2 = 0.007). We conclude that within the first 6 weeks of life, the infant microbiota undergoes substantial reorganization, which is primarily driven by body site and not by mode of delivery.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
Arumugam, M. et al. Enterotypes of the human gut microbiome. Nature 473, 174–180 (2011).
Aagaard, K. et al. A metagenomic approach to characterization of the vaginal microbiome signature in pregnancy. PLoS One 7, e36466 (2012).
Costello, E.K. et al. Bacterial community variation in human body habitats across space and time. Science 326, 1694–1697 (2009).
Turnbaugh, P.J. et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031 (2006).
Ravel, J. et al. Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. USA 108 (Suppl. 1), 4680–4687 (2011).
Ridaura, V.K. et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341, 1241214 (2013).
Qin, J. et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490, 55–60 (2012).
Morgan, X.C. et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 13, R79 (2012).
Schulz, M.D. et al. High-fat-diet-mediated dysbiosis promotes intestinal carcinogenesis independently of obesity. Nature 514, 508–512 (2014).
Gordon, H.A. & Pesti, L. The gnotobiotic animal as a tool in the study of host microbial relationships. Bacteriol. Rev. 35, 390–429 (1971).
Olszak, T. et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science 336, 489–493 (2012).
Wesemann, D.R. et al. Microbial colonization influences early B lineage development in the gut lamina propria. Nature 501, 112–115 (2013).
Gomez de Agüero, M. et al. The maternal microbiota drives early postnatal innate immune development. Science 351, 1296–1302 (2016).
Ivanov, I.I. et al. Induction of intestinal TH17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009).
Smith, P.M. et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341, 569–573 (2013).
Atarashi, K. et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331, 337–341 (2011).
Costello, E.K., Carlisle, E.M., Bik, E.M., Morowitz, M.J. & Relman, D.A. Microbiome assembly across multiple body sites in low-birthweight infants. mBio 4, e00782–13 (2013).
Feng, X.L., Xu, L., Guo, Y. & Ronsmans, C. Factors influencing rising cesarean section rates in China between 1988 and 2008. Bull. World Health Organ. 90, 30–39 (2012).
Osterman, M.J.K. & Martin, J.A. Trends in low-risk cesarean delivery in the United States, 1990–2013. Natl. Vital Stat. Rep. 63, 1–16 (2014).
Almqvist, C., Cnattingius, S., Lichtenstein, P. & Lundholm, C. The impact of birth mode of delivery on childhood asthma and allergic diseases—a sibling study. Clin. Exp. Allergy 42, 1369–1376 (2012).
Black, M., Bhattacharya, S., Philip, S., Norman, J.E. & McLernon, D.J. Planned repeat cesarean section at term and adverse childhood health outcomes: a record-linkage study. PLoS Med. 13, e1001973 (2016).
American College of Obstetricians and Gynecologists (College) & Society for Maternal–Fetal Medicine. Safe prevention of the primary cesarean delivery. Am. J. Obstet. Gynecol. 210, 179–193 (2014).
Dominguez-Bello, M.G. et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 107, 11971–11975 (2010).
Biasucci, G. et al. Mode of delivery affects the bacterial community in the newborn gut. Early Hum. Dev. 86 (Suppl. 1), 13–15 (2010).
Fallani, M. et al. Intestinal microbiota of 6-week-old infants across Europe: geographic influence beyond delivery mode, breast-feeding and antibiotics. J. Pediatr. Gastroenterol. Nutr. 51, 77–84 (2010).
Azad, M.B. et al. Gut microbiota of healthy Canadian infants: profiles by mode of delivery and infant diet at 4 months. CMAJ 185, 385–394 (2013).
Barber, E.L. et al. Indications contributing to the increasing cesarean delivery rate. Obstet. Gynecol. 118, 29–38 (2011).
David, L.A. et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014).
Koenig, J.E. et al. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. USA 108 (Suppl. 1), 4578–4585 (2011).
Jost, T., Lacroix, C., Braegger, C.P., Rochat, F. & Chassard, C. Vertical mother–neonate transfer of maternal gut bacteria via breastfeeding. Environ. Microbiol. 16, 2891–2904 (2014).
Azad, M. et al. Impact of maternal intrapartum antibiotics, method of birth and breastfeeding on gut microbiota during the first year of life: a prospective cohort study. BJOG Int. J. Obstet. Gynaecol. 123, 983–993 (2015).
Ardeshir, A. et al. Breast-fed and bottle-fed infant rhesus macaques develop distinct gut microbiotas and immune systems. Sci. Transl. Med. 6, 252ra120 (2014).
Dufrene, M. & Legendre, P. Species assemblages and indicator species: the need for a flexible asymmetrical approach. Ecol. Monogr. 67, 345–366 (1997).
Koren, O. et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell 150, 470–480 (2012).
La Rosa, P.S. et al. Patterned progression of bacterial populations in the premature infant gut. Proc. Natl. Acad. Sci. USA 111, 12522–12527 (2014).
Aagaard, K. et al. The placenta harbors a unique microbiome. Sci. Transl. Med. 6, 237ra65 (2014).
Antony, K.M. et al. The pre-term placental microbiome varies in association with excess maternal gestational weight gain. Am. J. Obstet. Gynecol. 212, 653.e1–653.e16 (2015).
Collado, M.C., Rautava, S., Aakko, J., Isolauri, E. & Salminen, S. Human gut colonization may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci. Rep. 6, 23129 (2016).
Bäckhed, F. et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17, 852 (2015).
Knights, D. et al. Bayesian community-wide culture-independent microbial source tracking. Nat. Methods 8, 761–763 (2011).
Penders, J. et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 118, 511–521 (2006).
Vatanen, T. et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell 165, 842–853 (2016).
Dominguez-Bello, M.G. et al. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat. Med. 22, 250–253 (2016).
Galley, J.D., Bailey, M., Kamp Dush, C., Schoppe-Sullivan, S. & Christian, L.M. Maternal obesity is associated with alterations in the gut microbiome in toddlers. PLoS One 9, e113026 (2014).
Mueller, N.T. et al. Birth-mode-dependent association between pre-pregnancy maternal weight status and the neonatal intestinal microbiome. Sci. Rep. 6, 23133 (2016).
Chu, D.M. et al. The early infant gut microbiome varies in association with a maternal high-fat diet. Genome Med. 8, 77 (2016).
Hu, J. et al. Diversified microbiota of meconium is affected by maternal diabetes status. PLoS One 8, e78257 (2013).
Ridlon, J.M., Kang, D.J., Hylemon, P.B. & Bajaj, J.S. Bile acids and the gut microbiome. Curr. Opin. Gastroenterol. 30, 332–338 (2014).
Kennedy, D.O. B vitamins and the brain: mechanisms, dose and efficacy—a review. Nutrients 8, 68 (2016).
Gibson, M.K. et al. Developmental dynamics of the preterm infant gut microbiota and antibiotic resistome. Nat. Microbiol. 1, 16024 (2016).
Bokulich, N.A. et al. Antibiotics, birth mode and diet shape microbiome maturation during early life. Sci. Transl. Med. 8, 343ra82 (2016).
DiGiulio, D.B. et al. Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: a molecular- and culture-based investigation. PLoS One 3, e3056 (2008).
Steel, J.H. et al. Bacteria and inflammatory cells in fetal membranes do not always cause preterm labor. Pediatr. Res. 57, 404–411 (2005).
Jiménez, E. et al. Isolation of commensal bacteria from umbilical cord blood of healthy neonates born by cesarean section. Curr. Microbiol. 51, 270–274 (2005).
Jiménez, E. et al. Is meconium from healthy newborns actually sterile? Res. Microbiol. 159, 187–193 (2008).
Ardissone, A.N. et al. Meconium microbiome analysis identifies bacteria correlated with premature birth. PLoS One 9, e90784 (2014).
Hansen, R. et al. First-pass meconium samples from healthy-term vaginally delivered neonates: an analysis of the microbiota. PLoS One 10, e0133320 (2015).
Fallani, M. et al. Determinants of the human infant intestinal microbiota after the introduction of first complementary foods in infant samples from five European centers. Microbiology 157, 1385–1392 (2011).
Yassour, M. et al. Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci. Transl. Med. 8, 343ra81 (2016).
La Rosa, P.S. et al. Hypothesis testing and power calculations for taxonomic-based human microbiome data. PLoS One 7, e52078 (2012).
Aagaard, K. et al. The Human Microbiome Project strategy for comprehensive sampling of the human microbiome and why it matters. FASEB J. 27, 1012–1022 (2013).
Caporaso, J.G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010).
Schmieder, R. & Edwards, R. Fast identification and removal of sequence contamination from genomic and metagenomic data sets. PLoS One 6, e17288 (2011).
Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010).
Haas, B.J. et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21, 494–504 (2011).
Cole, J.R. et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37, D141–D145 (2009).
DeSantis, T.Z. et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072 (2006).
Segata, N. et al. Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60 (2011).
Segata, N. et al. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat. Methods 9, 811–814 (2012).
Abubucker, S. et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput. Biol. 8, e1002358 (2012).
Le Chatelier, E. et al. Richness of human gut microbiome correlates with metabolic markers. Nature 500, 541–546 (2013).
Acknowledgements
The authors gratefully acknowledge the support of the NIH Director's New Innovator Award (DP2 DP21DP2OD001500; K.M. Aagaard), the NIH–NINR (NR014792-01; K.M. Aagaard), the NIH National Children's Study Formative Research (N01-HD-80020; K.M. Aagaard), the Burroughs Welcome Fund Preterm Birth Initiative (K.M. Aagaard), the March of Dimes Preterm Birth Research Initiative (K.M. Aagaard), the Baylor College of Medicine Medical Scientist Training Program (NIH NIGMS T32 GM007330; D.C. and K.M. Aagaard), the National Institute of General Medical Sciences (T32GM088129; D.M.C.), Baylor Research Advocates for Student Scientists (D.M.C.) and the Human Microbiome Project funded through the NIH Director's Common Fund at the National Institutes of Health (as part of NIH RoadMap 1.5; K.M. Aagaard). All sequencing and adaptation of protocols for WGS sequencing were performed by the Baylor College of Medicine Human Genome Sequencing Center (BCM–HGSC), which is funded by direct support from the National Human Genome Research Institute (NHGRI) at NIH (U54HG004973 (BCM); R. Gibbs, Principal Investigator). The authors also thank the staff members who were directly involved in clinical recruitment and specimen processing (M. Moller, B. Boggan, R. Benjamin, J. Chen, C. Cook and D. Racusin). The authors are grateful to M. Belfort, J. Versalovic, T. Savidge, R.A. Luna, D. Racusin, M. Suter and K. Meyer for critical review of the manuscript.
Author information
Authors and Affiliations
Contributions
D.M.C. and K.M. Aagaard designed and conceived the study; K.M. Aagaard and K.M. Antony assembled the cohort and developed the infrastructure to obtain swabs, samples and clinical metadata from all samples; K.M. Aagaard and K.M. Antony recruited and sampled all subjects; A.L.P., D.M.C. and M.D.S. prepared samples for sequencing of the gene encoding16S rRNA and for WGS sequencing; D.M.C., J.M. and K.M. Aagaard performed and supervised all analysis and statistical modeling; and D.M.C. and K.M. Aagaard wrote the manuscript, with contributions from J.M., A.L.P., K.M. Antony and M.D.S.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figures
Supplementary Figures 1–16 (PDF 5996 kb)
Supplementary Tables
Supplementary Tables 1–12 (XLSX 5234 kb)
Rights and permissions
About this article
Cite this article
Chu, D., Ma, J., Prince, A. et al. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat Med 23, 314–326 (2017). https://doi.org/10.1038/nm.4272
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.4272