Abstract
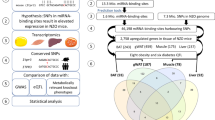
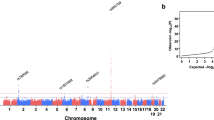
Genome-wide association studies (GWASs) have linked genes to various pathological traits. However, the potential contribution of regulatory noncoding RNAs, such as microRNAs (miRNAs), to a genetic predisposition to pathological conditions has remained unclear. We leveraged GWAS meta-analysis data from >188,000 individuals to identify 69 miRNAs in physical proximity to single-nucleotide polymorphisms (SNPs) associated with abnormal levels of circulating lipids. Several of these miRNAs (miR-128-1, miR-148a, miR-130b, and miR-301b) control the expression of key proteins involved in cholesterol-lipoprotein trafficking, such as the low-density lipoprotein (LDL) receptor (LDLR) and the ATP-binding cassette A1 (ABCA1) cholesterol transporter. Consistent with human liver expression data and genetic links to abnormal blood lipid levels, overexpression and antisense targeting of miR-128-1 or miR-148a in high-fat diet–fed C57BL/6J and Apoe-null mice resulted in altered hepatic expression of proteins involved in lipid trafficking and metabolism, and in modulated levels of circulating lipoprotein-cholesterol and triglycerides. Taken together, these findings support the notion that altered expression of miRNAs may contribute to abnormal blood lipid levels, predisposing individuals to human cardiometabolic disorders.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Cornier, M.A. et al. The metabolic syndrome. Endocr. Rev. 29, 777–822 (2008).
Go, A.S. et al. Heart disease and stroke statistics–2013 update: a report from the American Heart Association. Circulation 127, e6–e245 (2013).
Quiat, D. & Olson, E.N. MicroRNAs in cardiovascular disease: from pathogenesis to prevention and treatment. J. Clin. Invest. 123, 11–18 (2013).
Rottiers, V. & Naar, A.M. MicroRNAs in metabolism and metabolic disorders. Nat. Rev. Mol. Cell Biol. 13, 239–250 (2012).
Najafi-Shoushtari, S.H. et al. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science 328, 1566–1569 (2010).
Rayner, K.J. et al. Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J. Clin. Invest. 121, 2921–2931 (2011).
Marquart, T.J., Allen, R.M., Ory, D.S. & Baldan, A. miR-33 links SREBP-2 induction to repression of sterol transporters. Proc. Natl. Acad. Sci. USA 107, 12228–12232 (2010).
Horie, T. et al. MicroRNA-33 encoded by an intron of sterol regulatory element-binding protein 2 (Srebp2) regulates HDL in vivo. Proc. Natl. Acad. Sci. USA 107, 17321–17326 (2010).
Rayner, K.J. et al. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature 478, 404–407 (2011).
Goedeke, L., Aranda, J.F. & Fernandez-Hernando, C. microRNA regulation of lipoprotein metabolism. Curr. Opin. Lipidol. 25, 282–288 (2014).
Willer, C.J. et al. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 45, 1274–1283 (2013).
Merino, D.M., Ma, D.W. & Mutch, D.M. Genetic variation in lipid desaturases and its impact on the development of human disease. Lipids Health Dis. 9, 63 (2010).
Charles, M.A. & Kane, J.P. New molecular insights into CETP structure and function: a review. J. Lipid Res. 53, 1451–1458 (2012).
Lewis, B.P., Burge, C.B. & Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20 (2005).
Dennis, G. Jr. et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4, P3 (2003).
Ishibashi, S. et al. Hypercholesterolemia in low density lipoprotein receptor knockout mice and its reversal by adenovirus-mediated gene delivery. J. Clin. Invest. 92, 883–893 (1993).
Brown, M.S. & Goldstein, J.L. How LDL receptors influence cholesterol and atherosclerosis. Sci. Am. 251, 58–66 (1984).
Tall, A.R., Yvan-Charvet, L., Terasaka, N., Pagler, T. & Wang, N. HDL, ABC transporters, and cholesterol efflux: implications for the treatment of atherosclerosis. Cell Metab. 7, 365–375 (2008).
Navab, M., Reddy, S.T., Van Lenten, B.J. & Fogelman, A.M. HDL and cardiovascular disease: atherogenic and atheroprotective mechanisms. Nat. Rev. Cardiol. 8, 222–232 (2011).
Duffy, D. & Rader, D.J. Update on strategies to increase HDL quantity and function. Nat. Rev. Cardiol. 6, 455–463 (2009).
Sparks, J.D., Sparks, C.E. & Adeli, K. Selective hepatic insulin resistance, VLDL overproduction, and hypertriglyceridemia. Arterioscler. Thromb. Vasc. Biol. 32, 2104–2112 (2012).
Stein, S. & Matter, C.M. Protective roles of SIRT1 in atherosclerosis. Cell Cycle 10, 640–647 (2011).
Yoon, Y.S., Seo, W.Y., Lee, M.W., Kim, S.T. & Koo, S.H. Salt-inducible kinase regulates hepatic lipogenesis by controlling SREBP-1c phosphorylation. J. Biol. Chem. 284, 10446–10452 (2009).
Hardie, D.G. Organismal carbohydrate and lipid homeostasis. Cold Spring Harb. Perspect. Biol. 4 (2012).
Bonnefont, J.P. et al. Carnitine palmitoyltransferases 1 and 2: biochemical, molecular and medical aspects. Mol. Aspects Med. 25, 495–520 (2004).
Goldstein, J.L., DeBose-Boyd, R.A. & Brown, M.S. Protein sensors for membrane sterols. Cell 124, 35–46 (2006).
Goldstein, J.L. & Brown, M.S. The LDL receptor. Arterioscler. Thromb. Vasc. Biol. 29, 431–438 (2009).
Soutar, A.K. & Naoumova, R.P. Mechanisms of disease: genetic causes of familial hypercholesterolemia. Nat. Clin. Pract. Cardiovasc. Med. 4, 214–225 (2007).
Lu, Y.C. et al. ELAVL1 modulates transcriptome-wide miRNA binding in murine macrophages. Cell Rep. 9, 2330–2343 (2014).
Zhang, S.H., Reddick, R.L., Piedrahita, J.A. & Maeda, N. Spontaneous hypercholesterolemia and arterial lesions in mice lacking apolipoprotein E. Science 258, 468–471 (1992).
Nakashima, Y., Plump, A.S., Raines, E.W., Breslow, J.L. & Ross, R. ApoE-deficient mice develop lesions of all phases of atherosclerosis throughout the arterial tree. Arterioscler. Thromb. 14, 133–140 (1994).
Teslovich, T.M. et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466, 707–713 (2010).
Hatoum, I.J. et al. Melanocortin-4 receptor signaling is required for weight loss after gastric bypass surgery. J. Clin. Endocrinol. Metab. 97, E1023–E1031 (2012).
Greenawalt, D.M. et al. A survey of the genetics of stomach, liver, and adipose gene expression from a morbidly obese cohort. Genome Res. 21, 1008–1016 (2011).
Hatoum, I.J. et al. Weight loss after gastric bypass is associated with a variant at 15q26.1. Am. J. Hum. Genet. 92, 827–834 (2013).
Raymond, C.K., Roberts, B.S., Garrett-Engele, P., Lim, L.P. & Johnson, J.M. Simple, quantitative primer-extension PCR assay for direct monitoring of microRNAs and short-interfering RNAs. RNA 11, 1737–1744 (2005).
Bates, D. et al. Fitting linear mixed-effects models using lme4. arXiv, http://arxiv.org/abs/1406.5823 (2014).
Liu, E.Y., Li, M., Wang, W. & Li, Y. MaCH-admix: genotype imputation for admixed populations. Genet. Epidemiol. 37, 25–37 (2013).
Price, A.L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Shabalin, A.A. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics 28, 1353–1358 (2012).
Weischenfeldt, J. & Porse, B. Bone marrow-derived macrophages (BMM): isolation and applications. CSH Protoc 2008 doi:10.1101/pdb.prot5080 (2010).
Hafner, M. et al. Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods 44, 3–12 (2008).
Hafner, M. et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141, 129–141 (2010).
Corcoran, D.L. et al. PARalyzer: definition of RNA binding sites from PAR-CLIP short-read sequence data. Genome Biol. 12, R79 (2011).
Mattison, J.A. et al. Resveratrol prevents high fat/sucrose diet-induced central arterial wall inflammation and stiffening in nonhuman primates. Cell Metab. 20, 183–190 (2014).
Rayner, K.J. et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Science 328, 1570–1573 (2010).
Lillie, R.D. & Fulmer, H.M. Histopathologic Technique and Practical Histochemistry. (McGraw-Hill, 1976).
Ljungberg, O. & Tibblin, S. Peroperative fat staining of frozen sections in primary hyperparathyroidism. Am. J. Pathol. 95, 633–641 (1979).
Galabova, G. et al. Peptide-based anti-PCSK9 vaccines - an approach for long-term LDLc management. PLoS ONE 9, e114469 (2014).
Acknowledgements
This work was supported by the following grants: US National Institutes of Health (NIH) grants R21DK084459 and R01DK094184 (A.M.N); R37DK048873 and R01DK056626 to D.E.C.; K24DK078772 to R.T.C.; and R01HL107953 and R01HL106063 to C.F.-H. A.M.N. was supported by a scholar award from the Massachusetts General Hospital (MGH). R.E.G. was supported by an Established Investigator Award from the American Heart Association. C.F.-H. was supported by a Fondation Leducq Transatlantic Network of Excellence in Cardiovascular Research Award. T.H. acknowledges the support of NIH grant R01HL49094 and a Fondation Leducq Transatlantic Network of Excellence in Cardiovascular Research Award. N.S. and J.S.T. were supported by the Intramural Research Program of the US National Institute of Allergy and Infectious Diseases. C.M.R. was supported by the American Heart Association SDG Grant 15SDG23000025. A.W. was supported by a fellowship from the MGH Executive Committee on Research. We thank C. Molony of Merck Research Laboratories for help with genotyping data quality assessment. We thank the Harvard Medical School Neurobiology Department and the Neurobiology Imaging Facility for consultation and instrument availability that supported this work. This facility is supported in part by the Neural Imaging Center as part of a National Institutes on Neurological Disorders and Stroke P30 Core Center grant no. NS072030, and by the Harvard Digestive Diseases Center (P30 DK034854).
Author information
Authors and Affiliations
Contributions
S.H.N.-S. and A.M.N. conceived and carried out the initial miR-128-1 studies that provided support for the expanded project. A.W. and A.M.N. conceived and designed the expanded, published studies, interpreted the data, and wrote the manuscript, which was commented on by all authors. A.W., S.H.N.-S., L.W., S.S., Y. Li, F.K., N.P., D.E.C. and R.E.G. performed experiments and analyzed data in Apoe deficient mice and in C57BL/6 mice. L.G., C.M.R. and C.F.-H. quantified miRNAs in Rhesus monkeys and mice fed with different diets and performed efflux experiments in mouse peritoneal macrophages. N.S., Y. Lu, J.S.T., E.S., R.T., I.H., P.C.S. and L.M.K. contributed to the human liver miR-QTL analysis. Y.-C.L. and T.H. performed the Ago2 PAR-CLIP analysis in BMDMs. A.S.d. and R.T.C. performed the liver histology analysis. V.V. injected the lentivirus in C57BL/6 mice. J.C.B. and J.W. performed DNA microarray analysis from mouse liver samples prepared by A.W. S.K. and A.B. analyzed the GWAS data. R.d.C. carried out the non-human primate studies.
Corresponding author
Ethics declarations
Competing interests
A.M.N., A.W. and S.H.N.-S. have issued and pending patents (US Patent nos. 9,045,749 and US 61/865,327) on microRNA-targeting therapeutics for the treatment of cardiometabolic diseases.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 (PDF 3650 kb)
Supplementary Table 1
69 miRNAs are associated with abnormal levels of total cholesterol (TC), LDL-C, HDL-C, and TAGs. (XLSX 65 kb)
Supplementary Table 2
Gene Ontology analysis of miRNA target genes. (XLSX 37 kb)
Supplementary Table 3
List of SNPs associated with total cholesterol, LDL-C, HDL- C and triglycerides in the miR-128-1, miR-148a, miR-130b and miR-301b loci. (XLSX 121 kb)
Supplementary Table 4
Selected metabolism genes predicted to be targeted by miR- 128-1, miR-148a, miR-130b and miR-301b. (XLSX 46 kb)
Supplementary Table 5
Liver cis miR-QTL data. (XLSX 1002 kb)
Supplementary Table 6
Expression changes of predicted miR-128-1 targets in the livers of mice treated with antimiR-128-1 versus control scrambled LNA antimiR over 5 days. (XLSX 91 kb)
Rights and permissions
About this article
Cite this article
Wagschal, A., Najafi-Shoushtari, S., Wang, L. et al. Genome-wide identification of microRNAs regulating cholesterol and triglyceride homeostasis. Nat Med 21, 1290–1297 (2015). https://doi.org/10.1038/nm.3980
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.3980
This article is cited by
-
MicroRNAs and Cardiovascular Disease Risk
Current Cardiology Reports (2024)
-
Lipid metabolism-related miRNAs with potential diagnostic roles in prostate cancer
Lipids in Health and Disease (2023)
-
Functional significance of cholesterol metabolism in cancer: from threat to treatment
Experimental & Molecular Medicine (2023)
-
Systematic characterization of seed overlap microRNA cotargeting associated with lupus pathogenesis
BMC Biology (2022)
-
Aberrant expression of HDL-bound microRNA induced by a high-fat diet in a pig model: implications in the pathogenesis of dyslipidaemia
BMC Cardiovascular Disorders (2021)