Abstract
Central memory T (TCM) cells in lymph nodes (LNs) and resident memory T (TRM) cells in peripheral tissues have distinct roles in protective immunity1,2,3,4,5. Both are generated after primary infections, but their clonal origins have been unclear. To address this question, we immunized mice through the skin with a protein antigen, a chemical hapten, or a non-replicating poxvirus. We then analyzed antigen-activated T cells from different tissues using high-throughput sequencing (HTS) of the gene encoding the T cell receptor (TCR) β-chain (Trb, also known as Tcrb) using CDR3 sequences to simultaneously track thousands of unique T cells6. For every abundant TRM cell clone generated in the skin, an abundant TCM cell clone bearing the identical TCR was present in the LNs. Thus, antigen-reactive skin TRM and LN TCM cell clones were derived from a common naive T cell precursor after skin immunization, generating overlapping TCR repertoires. Although they bore the same TCR, TRM cells mediated rapid contact hypersensitivity7 responses, whereas TCM cells mediated delayed and attenuated responses. Studies in human subjects confirmed the generation of skin TRM cells in allergic contact dermatitis. Thus, immunization through skin simultaneously generates skin TRM and LN TCM cells in similar numbers from the same naive T cells.
Similar content being viewed by others
Main
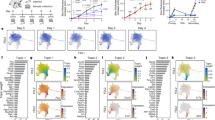
TRM cells are known to be important in protective immunity, but their origin and relationship to other memory T cell subsets remain obscure. We previously demonstrated that both TRM and TCM cells were generated after vaccinia skin infection8,9, but the lineage commitment of individual naive T cells to TRM or TCM cells in vivo was not addressed. Some studies of endogenous naive T cells in vivo have suggested a one-cell, one-fate paradigm5,10,11,12,13. One report demonstrated that differentiation of individual naive CD4+ T cells into effector (Teff) or follicular helper (TFH) T cells was determined by TCR affinity for antigen13, whereas another report showed that microenvironmental factors influenced the differentiation of individual naive CD8+ T cells into TEM or TCM cells12. In contrast, other studies using naive TCR transgenic CD8+ T cells demonstrated that commitment of single T cells to either the TEM or the TCM lineage was stochastic14,15, a finding that is inconsistent with the one-cell, one-fate paradigm. To explore the origin of TCM and TRM cells further, we used three different antigens to immunize mouse skin: ovalbumin (OVA) with adjuvant cholera toxin (CT), a contact sensitizer (dinitrofluorobenzene, DNFB), and the poxvirus modified Vaccinia Ankara (MVA) (Supplementary Fig. 1). In OVA + CT experiments, OT-I transgenic T cells, which bear a TCR specific to OVA257–264, were injected into mice before immunization as an internal control (Supplementary Fig. 1). After each immunization, acute inflammation had resolved completely by day 35, and resting CD4+ and CD8+ T cells remained at the antigen treated site8,16 (Supplementary Figs. 2 and 3). We extracted DNA from skin and LN of individual mice before and at different time points after immunization and performed HTS of Trb as previously described6,17,18. After 35 d, expanded T cell clones that were undetectable before immunization were considered antigen-specific, and the most abundant memory T cell clones (detectable at ≥5 copies/400 ng of DNA at the site of antigen encounter) were studied further. After OVA + CT application, T cell clones present in both immunized ear and in distant unchallenged skin were represented as blue dots corresponding to unique CDR3 sequences (Fig. 1a), consistent with our previous finding that skin infection generates antigen-specific TRM cell clones in both treated and distant skin8. The six expanded T cells analyzed (Fig. 1a, blue box) were undetectable before immunization in skin and LN (except for the OT-1 CDR3 in LN) but abundant in skin and LN after immunization (Fig. 1b). The data are expressed as 'clone frequency', and although all clones were present in both skin and LN after immunization, the value for each clone was lower in LN than in skin (Fig. 1b), as the absolute number of T cells is much higher in LN than in skin. Therefore, we measured the total number of T cells of a particular clone in skin versus LN by measuring the number of CDR3-identical T cells per 400 ng of genomic DNA (gDNA). By this analysis, TRM cells in skin and TCM cells in LN with the same CDR3 sequences were present in comparable abundance after OVA + CT immunization (Supplementary Fig. 1). Of the 15 most abundant clones found in ear and tail skin, nearly all were also present and abundant in both distant and draining LNs (Fig. 1c). Comparable results were obtained after DNFB (Fig. 1d–f) and MVA (Fig. 1g–i) exposure. For all three types of immunization, treated and distant skin (TRM cells) as well as draining and distant LNs contained common expanded memory T cell clones that were not detectable before immunization. Memory T cells enter distant LNs directly from the blood through high endothelial venules rather than through afferent lymphatics; thus, memory T cell clones in distant LNs were considered TCM cells. In all mice from all groups combined, 85–93% of TRM cell clones that were expanded at both skin sites were mirrored by CDR3-identical LN TCM cell clones in similar abundance (n = 41; Fig. 1j). We concluded that these TRM and TCM cells were derived from common naive T cell precursors.
(a–c) Dot plots of the frequency (number of a given sequence divided by the total number of sequences observed in the given sample) of TCR-β CDR3 amino acid (AA) sequences shared in antigen-exposed (ear, green vertical axis) and distant (tail, red horizontal axis) skin 6 weeks after OVA + CT immunization (a), DNFB sensitization (b), and MVA skin scarification (c). Each blue dot represents a T cell clone with a unique CDR3 sequence, and the most abundant expanded clones are enclosed in the blue rectangle. CASSRANYEQYF is the OT-I TCRVβ CDR3 sequence. n = 3 mice per antigen. (d–f) Tracking of selected abundant TCR-β CDR3 sequences after OVA + CT (d), DNFB (e), and MVA (f). n = 3 mice per antigen. I, inguinal LN; T, tail skin; E, ear skin; D, draining LN (ear draining). Data for CDR3 sequences (TCR percentage) are shown as the percentage (specific/total) of CDR3 sequences from T cell clones present at n ≥ 5 copies in skin assessed in other tissues. (g–i) Unique expanded clones after OVA + CT (g), DNFB (h), and MVA (i) in ear and tail skin and the corresponding clones present in draining and distant LNs. Each color represents a unique clone. The average clone size in each compartment is shown in the colored circle. (j) Total number of T cell clones (n ≥ 5 per clone, horizontal bar graphs) and mean clone size (colored circle) in the tissues of all mice combined, n = 9 mice.
We next studied T cell responses following multiple applications of the hapten DFNB to the same skin site (Fig. 2a). We performed tissue harvest and HTS at day 91 after the initial sensitization (Fig. 2b–d). We compared the abundance of T cells in particular VJ groups between three skin sites—the tail (no DNFB), the right ear (DNFB × 2), and the left ear (DNFB × 6)—and the LNs draining these sites (Fig. 2b). The expansion of individual VJ groups as a function of DNFB exposure was greater in the skin than the LNs. In terms of V13.3/J2.2 T cell clones, three dominant clones (clones 1–3) were expanded in both skin and LN at day 91 after sensitization for all three DNFB exposure conditions. These same three abundant V13.3/J2.2 clones were consistently present in skin and LN at this time for all conditions (Fig. 2d). To rule out possible peripheral blood contamination of our tissue samples, we examined blood T cells by HTS of the Trb gene at the same time point (Supplementary Fig. 2). We found that nine of the ten most abundant T cell clones in blood were undetectable in skin and LN (Supplementary Fig. 2a); conversely, the most abundant clones in skin and LN were either absent or rare in blood (Supplementary Fig. 2a). Thus, our results cannot be attributed to peripheral blood contamination. After six DNFB applications, the most abundant clones in skin were 7–17× more numerous in skin than in LN (Fig. 2e,f). We conclude that multiple exposures of the same antigen to skin lead to preferential expansion of skin TRM cells. Increased numbers of both CD4+ and CD8+ T cells (Supplementary Fig. 3) expressing a TRM cell phenotype were present in skin (Supplementary Fig. 4).
(a) Temporal scheme of the repetitive DNFB sensitizations and challenges. n = 3 mice for each condition, all experiments performed 2 or 3 times, representative data are shown. (b) Frequency of respective β chain V and J family T cell populations in the skin and LN of a repetitively immunized mouse at day 91. The x-axis shows the TCR Vβ subtype (described in detail in Online Methods), the y-axis shows their frequency, and the z-axis shows the TCR Jβ subtype. Top graphs show the Vβ, Jβ repertoire in different skin areas exposed to DFNB; bottom graphs show the same but in the respective draining LN (inguinal, right and left submandibular LNs for tail, right and left ear, respectively). n = 3 mice. (c) At left, pie chart of the frequency of the highly expanded V13.3/J2.2 clones in mice sensitized, boosted and challenged with DNFB, both in the left ear (skin) and left cervical (draining, d) LN. At right, pie chart of the three most abundant individual TCR clones (unique CDR3 sequences) and all other minor clones comprising this specific VJ subgroup. n = 3 mice. (d) Clone frequency of the three most abundant TCR clones from c in skin and LN after zero, two, and six DNFB exposures. (e) The single most abundant T cell clone in skin exposed zero, two, and six times to DNFB in three mice, expressed as T cells/400 ng of gDNA. The copy number of the same clone in LN is shown in parentheses (in the 6 exposures circles). (f) Number of highly expanded clones present at ≥10 copies per 400 ng of tissue in the skin and LN of the immunized mice, showing skin-specific (TRM cell) increase. n = 3 mice.
We next joined DNFB-sensitized mice to naive mice by parabiotic surgery (Fig. 3a). After 4 weeks of parabiosis followed by separation for 2 weeks, multiple shared clones were identified in the LNs of the naive and immunized parabionts, confirming free transfer of TCM cells between parabiotic mice (Supplementary Fig. 5). While a very small number of shared clones appeared to be shared in the skin after parabiosis and separation (Supplementary Fig. 5), this difference was not statistically significant (P = 0.1). These data demonstrate by HTS of the Trb gene that TCM cells are freely migratory and TRM cells in skin are sedentary. Despite the rapid equilibration of T cells between LNs in our parabionts8, there was low absolute concordance of T cells (% of total unique CDR3 sequences shared) between these LNs (∼3%). To explore this further, we performed additional experiments and determined that overlap of total unique CDR3 sequences between two different LNs in the same healthy 6-week-old mouse was actually very low (roughly 1%, Supplementary Fig. 6). This number did not increase appreciably by 4 weeks after DNFB sensitization. In the same mouse, overlap of unique CDR3 sequences between skin sites was 5% before and 11% after DNFB sensitization (Supplementary Fig. 6). However, when expressed as the overlap of total CDR3 sequences in skin (accounting for the abundance of certain clones), the values were 20% before and 50% after DNFB sensitization, consistent with results shown in Figure 2. The overlap of total CDR3 sequences in LNs rose from 5% before to 7% after DNFB (Supplementary Fig. 6), emphasizing that even though expanded memory T cell clones are detectable in LNs, they are greatly outnumbered by naive T cells. In contrast, skin contained only memory T cells.
(a) Diagram of the subsequent experiments. n = 3 pairs of parabiotic mice; 3 naive and 3 sensitized non-parabiotic controls. Experiments were performed 2–4 times. (b) Ear-swelling kinetics in sensitized and naive parabiotic mice, with sensitized and naive non-parabiotic mice as controls. *P = 0.001, §P = 0.005; n = 3 per group. (c) Same experiment as in b, except mice were pretreated with FTY-720. *P = 0.002, §P = 0.002; n = 3 per group. (d) Comparison of ear-swelling kinetics after DNFB challenge at the initial site (local, left ear, L) of immunization and at a distant site (dist., right ear, R) in parabiotic sensitized mice. In parabiotic naive mice, the left and right ears were measured after DNFB challenge. *P = 0.001, §P = 0.01, θP = 0.02; n = 5 mice per group. (e) Kinetics of left and right ear swelling after DNFB challenge in parabiotic naive mice, with or without FTY-720 treatment. *P = 0.001, §P = 0.001; n = 5 mice per group. (f) Left ear swelling after the final (second) DNFB challenge in both naive and sensitized parabiotic mice, treated as shown in a, with non-parabiotic naive and sensitized mice as controls. §P = 0.04; θP = 0.001; n = 5 mice per group. (g) Left (local) versus right (distant) ear swelling after the final DNFB challenge in both naive and sensitized parabiotic mice. *P = 0.04, n = 3 per group. (h) Ratio (%) of left versus right maximal ear swelling after the second DNFB challenge in sensitized and naive parabiotic mice. P = 0.029, n = 3 per group. All results show mean ± s.d. Statistical analysis was done using two-way ANOVA with Holm–Bonferroni post hoc analysis where appropriate, or one-way ANOVA only. P > 0.05 was considered not significant (NS).
Our data show that skin TRM and LN TCM cells generated by the same immunization share identical Trb CDR3 sequences. We used parabiotic mice8 and DNFB sensitization (Fig. 3) to examine the relative contributions of TRM and TCM cells to skin contact hypersensitivity (CHS), a mouse model of human allergic contact dermatitis (ACD)7,19,20. Previously sensitized parabiotic mice manifested robust ear swelling 24 h after DNFB challenge (Fig. 3b), consistent with the kinetics of a TRM cell response. Unsensitized (naive) parabiotic mice (that contain DNFB-specific TCM cells but not skin TRM cells) did not show increased ear swelling at 24 h, but developed a delayed ear-swelling response 5 d after challenge (Fig. 3b). Pretreatment of these unsensitized parabiotic mice with FTY-720, a sphingosine-1-phosphate receptor inhibitor (Supplementary Fig. 7), abrogated this delayed ear-swelling response (Fig. 3c), consistent with a requirement for TCM cell migration from LNs to induce inflammation. In contrast, FTY-720 had no effect on the rapid ear-swelling response to DNFB challenge seen in the sensitized parabiotic mice (Fig. 3c), confirming that this response was independent of T cell recruitment or migration9,21. These data are consistent with the rapid kinetics and durability of the TRM cell response to antigen, in contrast to the TCM cell response specific for the same antigen. We obtained complementary results in adoptive transfer studies at these time points (Supplementary Figs. 7–9).
When we challenged the unsensitized ear of the sensitized parabiotic mice with DNFB, we observed rapid ear swelling (24 h), though it was lower in magnitude than the (previously) sensitized ear (Fig. 3d). This is consistent with skin antigen challenge leading to a global seeding of skin with antigen specific TRM cells. In contrast, naive unsensitized parabiotic mice showed no rapid ear swelling (24 h) in either ear (Fig. 3d,h). Delayed ear swelling in both ears of these mice was observed at day 5 (Fig. 3d,h), which was abrogated by pretreatment with FTY-720, confirming that the delayed response was mediated by TCM cells (Fig. 3e). However, 4 weeks later, when these same mice were subsequently rechallenged on the sensitized ear, rapid ear swelling occurred at 24 h (Fig. 3f,g). This is consistent with de novo generation of TRM cells in these mice.
We next asked if TRM cells could be observed in the skin of human subjects with ACD. We sensitized healthy individuals (n = 11, 8 males and 3 females, ages 29–58) to the contact allergen diphenylcyclopropenone (DPCP) at day –14 on arm skin, and then challenged them with DPCP on left thigh skin and placebo on right thigh skin at day 0 (ref. 22 and Fig. 4a). We took biopsies of the challenged leg skin sites at day 3, 14, and month 4 after challenge (placebo-treated skin biopsies were only taken at day 3), and performed photographic and microscopic analysis. Clinically and microscopically, DPCP-challenged skin was inflamed at 3 and 14 d, but this had resolved by 4 months after challenge (Fig. 4b). For three of these individuals (selected randomly), we extracted DNA from skin biopsies and performed HTS of the TRB gene. We compared clones from placebo-challenged skin (presumably generated by sensitization) to those from DPCP-challenged skin biopsied at day 3 and identified a number of shared clones (Fig. 4c). We also identified five representative clones (1–5, red box, Fig. 4c) from skin biopsied day 14 and month 4 after challenge (Fig. 4d). The absolute number of each of these five T cell clones increased over time and remained high at 4 months (Fig. 4e), closely following the pattern and kinetics of antigen-specific TRM cell clones from our mouse studies. Additional T cell clones were present in placebo-treated skin, but these other clones either did not change or decreased in number over the course of the experiment (clones a–d, Supplementary Fig. 10), and others did not appear until 14 d after challenge (Supplementary Fig. 10), after which point their abundance decreased. These clones did not fit the criteria of TRM cells, and are consistent with a recent report showing that only a subset of T cells in human skin are authentic TRM cells23.
(a) Schematic of DPCP sensitization and sample recovery. (b) Clinical photos, H&E histology and CD3+ T cell staining of the skin of subjects at day 3 (both placebo and DPCP challenged), 14 (DPCP challenged) and 120 (DPCP challenged). Data representative of a total of 11 subjects entered in the study. (c,d) Dot plots of the frequency (number of a given sequence divided by the total number of sequences observed in the given sample) of TCR-β CDR3 sequences shared in non-exposed or DPCP-exposed skin from a representative individual. (c) Clones in a 3 d-post-DPCP challenge site skin (green, vertical axis) versus those in a placebo-challenged skin site at the same time point (red, horizontal axis). Clones present in both sites are shown as blue dots. Five clones are identified (1–5) for subsequent reference. (d) Clones present 14 d (green, vertical axis) versus 4 months (red, horizontal axis) in skin after DPCP challenge. The same five clones (labeled and enclosed in boxes) are identified as being present in both samples. (e) Quantification of the T cell clone size observed in c and d over time. n = 3 out of 11 subjects; F, 48 years old; M, 52 years old; M, 55 years old; biopsies were subjected to HTS; data from one representative patient is shown; each line represents one clone (of five representative clones mentioned in c and d).
In this study, we used HTS of the Trb gene to explore the generation, localization, and function of TRM and TCM cells in response to three types of skin immunization: a contact sensitizer hapten, a protein, and a virus. This approach allowed comprehensive characterization of the endogenous αβ T cell repertoires in normal nontransgenic mice24. After all immunizations, we observed the generation of comparable numbers of both TRM and TCM cell clones, residing in distinct anatomic compartments, from unique naive T cell precursors. Although one previous report showed that TRM cells were derived from killer cell lectin–like receptor G1 (KLRG1)lo precursors, the development of TRM cells from individual naive T cells has, to our knowledge, not been directly studied before25. In our study, the tissue TRM cell compartment provided rapid, tissue-specific responses to antigenic challenge, while the TCM cell compartment was slower to manifest effector function but could supplement the TRM cell compartment after antigen challenge. Even though analysis of Trb CDR3 sequences by HTS can yield false negatives, false positives are generally not observed (as this would require the observation of a very specific CDR3 sequence that is not genuinely present), suggesting that the 93% overlap at the level of common unique CDR3 sequences (Fig. 1h) may underestimate the true concordance between skin TRM and LN TCM cells. These data are consistent with the existence of a common naive T cell precursor of TRM and TCM cells, and therefore do not support the hypothesis that differentiation is determined by variables such as T cell receptor affinity or signaling strength5,10,11,12,13 Repetitive antigenic challenges to skin led to preferential expansion of skin TRM cells. We further demonstrated that TRM cells mediate CHS, the mouse counterpart of human ACD, an acquired human disease that has limited effective therapies and a high economic impact7. Our data from studies on ACD in human subjects using HTS of the TRB gene support our data from mouse studies. That ACD is mediated by TRM cells helps to explain the recalcitrant, recurrent, and site-specific nature of this disease and opens up new opportunities for therapies. By extension, we believe that many human immunologic diseases that recur episodically in barrier tissues (for example, psoriasis and inflammatory bowel disease) may be mediated by TRM cells. In summary, our observations suggest that after skin immunization, the immune system distributes TCR-identical, antigen-reactive T cells to both LNs (as TCM cells) and the peripheral tissues (as TRM cells), thus generating two compartments of memory T cells with identical antigen specificity but different effector properties.
Methods
Study subjects and skin samples.
Skin biopsies of DPCP-challenge reactions and placebo-treated sites were obtained from 11 volunteers (ages 29–58, 8 males and 3 females) under a protocol approved by The Rockefeller University's institutional review board22. We obtained written informed consent from all subjects and the study adhered to the Declaration of Helsinki principles. Consent to publish was obtained for all subjects who were photographed. Immunization with DPCP (Hapten Pharmaceuticals), recall and sampling have been described before22. Briefly, we sensitized healthy individuals to DPCP and 2 weeks later challenged them with two applications (0.04%) of DPCP on their left thigh and two placebo applications on their right thigh. We took skin biopsies at day 3, 14 and month 4 after challenge, and stored them at –80 °C until processing. For three subjects (one female, two males, age range 46–55, selected randomly), HTS was performed on all biopsy samples.
Animal experiments.
We purchased C57BL/6 mice (5–7 weeks of age, females, n = 302 total) from The Jackson Laboratory, Bar Harbor, ME. Purchased mice had at least 1 week of acclimation in the animal facility of Harvard Institutes of Medicine at Harvard Medical School, before the initiation of any experiment. We bred and housed C57BL/6J Thy1.1 Rag1−/− Tg(TcraTcrb)1100Mjb/J (better known as OT-I) mice at the same facility. We performed all animal experiments in accordance with the guidelines set out by the Center for Animal Resources and Comparative Medicine at Harvard Medical School. For all experiments, animals were randomized into the different experimental groups by the animal facility staff, so that the investigator did not select groups based on size or appearance, but the investigator was not blinded once the experiments were initiated. Groups of three mice for naive or simple DNFB-positive ear-swelling control experiments were sufficient as ear swelling is highly reproducible (pooled naive of positive control in this paper, 36 mice). For other conditions, we chose groups of five mice because prior publications had identified this as the correct number in regard to reaching statistical power with the minimal number of animals. The sole exclusion criterion was death of the subject animal (one occurrence of unknown cause, five others due to complications linked with the surgical procedure).
Antigen challenge to skin.
We sensitized C57BL/6 mice by topical applications of 20 μl of 0.25% DNFB diluted in acetone:olive oil(aOO) (3:1 v/v) at the indicated time points (day 0 and 1, or 0, 1 and 7). Challenged was performed at the indicated time points with 20 μl of DNFB–aOO (0.25%). We measured ear thickness with a digital thickness gauge (Mitutoyo) before and after DNFB challenge. For OVA + CT immunization, mice were immunized epicutaneously as previously described8. Briefly, we used scotch tape (3M) to gently remove the cornified layer of ear skin, and then treated the skin with acetone and cholera toxin adjuvant (List Biological Labs) before administrating chicken ovalbumin + cholera toxin. For OT-I experiments, we transferred 1 × 104–1 × 106 OT-I splenocytes intravenously into recipient mice 24 h before immunization of LN dissection. We performed epicutaneous infection by scarification as previously described9. Every experiment was performed at least twice (and often multiple times). We used fingolimod (FTY-720, Sigma) diluted in PBS and injected it intraperitoneally (i.p.) at 1 μg per g of mouse body weight: for example, typically 25 μg/mouse i.p. daily for 7 d, beginning 2 d before the DNFB challenge.
Parabiotic surgery.
We performed parabiosis surgery as previously described8. Briefly, we shaved the flank of sex- and age-matched mice 1 d before surgery. We then anesthetized them to full muscle relaxation with ketamine and xylazine (10 mg/g) i.p. After skin disinfection (betadine solution and 70% ethanol, 3×), we made two matching skin incisions from the olecranon to the knee joint of each mouse pair. We then bound the elbow and knee joints by a single 5-0 silk suture and closed the dorsal and ventral skins with staples and 5-0 silk suture. Betadine solution was used to cover the full length of the dorsal and ventral incision. We kept the mice on heating pads and continuously monitored them until recovery from anesthesia and surgery. We used 2.5 mg/g flunixin subcutaneously for analgesia every 12 h for 48 h after the operation. After an interval of the indicated weeks, parabiotic mice were surgically separated by a reversal of the above procedure for subsequent experiments.
Lymphadenectomy.
Briefly, we shaved mice on the flank and belly 1 d before surgery. We anesthetized the mice to full muscle relaxation with ketamine and xylazine (10 mg/g) i.p. After skin disinfection (betadine solution and 70% ethanol, 3×), we performed a 1 cm skin incision on a longitudinal line 0.5 cm medial to the mammary gland. We everted the lateral skin to reveal the inguinal lymph node, which we then excised. We closed the incision with two surgical clips, disinfected with betadine 1%, kept the mice on heating pads and continuously monitored them until recovery. We used 2.5 mg/g flunixin subcutaneously for analgesia every 12 h for 24 h.
Viruses and infections.
We obtained recombinant MVA from B. Moss (US National Institutes of Health) and grew them on DF-1 cells. Viral titers were determined using CV-1 cells and 2 × 106 p.f.u. of MVA used for epicutaneous infection by skin scarification, as previously described8.
Preparation of cell suspensions.
We harvested lymph nodes and spleen and passed them through a 70-mm nylon cell strainer to prepare cell suspensions, in which we lysed red blood cells with hypotonic solution (Gibco). For skin tissue, we removed hair, chopped skin into small fragments and incubated them in Hank's balanced salt solution (Gibco) supplemented with 1 mg/ml collagenase A (Pharmingen) and 40 mg/ml DNase I (Roche) at 37 °C for 30 min. After filtering through a 70-mm nylon cell strainer, we collected the cells and washed them thoroughly with cold PBS before staining.
Antibodies and flow cytometry.
We obtained the following anti-mouse antibodies from BD Pharmingen: CD8a (53-6.7), CD4 (L3T4), CD45.1 (A20), CD44 (IM7), CD62L (MEL-14), CD69 (H1.2F3), CD103 (M290), CD122 (TM-Beta 1). We used all antibodies at a dilution of 1:200. We purchased fluorescence-conjugated anti-mouse CD127 (A7R34) from eBioscience. To examine E- or P-selectin ligand expression, we incubated cells with rmE-Selectin/Fc Chimera or rmP-Selectin/Fc Chimera (R&D Systems) in conjunction with APC-conjugated F(ab9)2 fragments of goat anti-human IgG F(c) antibody (cat. 109-136-098, Jackson ImmunoResearch). We analyzed data on a FACSCanto flow cytometer using FACSDiva software.
Isolation of gDNA and TRB high-throughput sequencing.
We extracted DNA from 22–24 mg of skin (ear or de-boned tail skin), vagina, lung, gut or 6–12 mg of LN using Qiagen's DNeasy mini-columns according to the manufacturer's instruction and stored it at −20 °C. We then shipped it on dry ice to Adaptive Biotechnologies. All TCR-β characterization was performed by Adaptive Biotechnologies using the ImmunoSeq TCR-β 'survey level' mouse assay. To address amplification bias in the multiplex PCR, we divided a single C57Bl/6 mouse thymus into 12 sections and ran each thymus section independently through HTS. Taking CDR3 sequences found in only one sample (which produces a population made up almost exclusively of CDR3 sequences represented by a single T cell in the input material), we calculated the read coverage (compared to average) associated with each V and J gene segment and used these factors to normalize sequencing output. To estimate the number of T cells bearing each rearrangement, we began with the same population of CDR3 sequences as above and calculated the mean number of sequencing reads obtained per T cell of input in this population (after normalizing for amplification bias). Next we divided the total sequencing output of each of the 12 thymus samples by this mean coverage to estimate the number of T cells present in each of the 12 subsections. These data were used to model the relationship between the number of unique CDR3 sequences obtained and the total number of T cells sequenced. For the 12 thymus sections, these values were highly correlated (total T cells = 1.22 * unique CDR3 rearrangements; r2 = 0.98). This relationship depends on the clonal structure of the T cell population but should only depart from our model in the case of highly clonal T cell populations. In each subsequent sample this relationship was used to estimate the total number of T cells assayed and thus the mean coverage per input T cell. Finally, the number of T cells bearing each unique CDR3 sequence was estimated as number of sequencing reads obtained/estimated mean coverage. OT-I TCR tracking was based on the OT-I Vβ5.2/Dβ2/Jβ2.6 sequence 5-TACTTCTGTGCCAGCTCTCGGGCCAATTATGAACAGTACTTCGGTCCCGGCA-CCAGGCT-3′ (YFCASSRANYEQYFGPGTR). TCR-β characterization of human samples (n = 3 subjects) was performed using the Adaptive Biotechnologies ImmunoSeq human TCR-β assay, survey level. All raw data can be accessed at http://adaptivebiotech.com/papers/nmed69531a.
VDJ family lists corresponding to graphs in Figure 2b.
The analyzed mouse TCR subfamilies are described here. They are portrayed in this order in Figure 2b, from left to right for both the x- and y-axis. TCR-BV1-0, -BV2-0, -BV3-0, -BV4-0, -BV5-0, -BV6-0, -BV7-0, -BV8-0, -BV9-0, -BV10-0, -BV12-1, -BV12-2, -BV12-3, -BV13-1, -BV13-2, -BV13-3, -BV14-0, -BV15-0, -BV16-0, -BV17-0, -BV19-0, -BV20-0, -BV21-0, -BV22-0, -BV23-0, -BV24-0, -BV26-0, -BV27-0, -BV28-0, -BV29-0, -BV30-0, -BV31-0. TCR-BJ1-1, -BJ1-2, -BJ1-3, -BJ1-4, -BJ1-5, -BJ1-6, -BJ1-7, -BJ2-1, -BJ2-2, -BJ2-3, -BJ2-4, -BJ2-5, -BJ2-7, -BJ1-5*1, -BJ1-5*2, -BJ1-5*3 (the last three use three different set of primers for the same J1-5 subtype). Human TCR subfamilies analyzed were TCR-BV01-01, BV02-01, BV04-01, -BV04-02, -BV04-03, -BV05-01, -BV05-03, -BV05-04, -BV05-05, -BV05-06, -BV05-08, -BV06-01, -BV06-04, -BV06-05, -BV06-06, -BV06-07, -BV06-08, -BV06-09, -BV07-02, -BV07-03, -BV07-04, -BV07-06, -BV07-07, -BV07-08, -BV07-09, -BV09-01, -BV10-01, -BV10-02, -BV10-03, -BV11-01, -BV11-02, -BV11-03, -BV12-01, -BV12-02, -BV12-05, -BV13-01, -BV14-01, -BV15-01, -BV16-01, -BV18-01, -BV19-01, -BV20-01, -BV21-01, -BV22-01, -BV23-01, -BV25-01, -BV27-01, -BV28-01, -BV29-01, -BV30-01 and TCR-BJ01-01, -BJ01-02, -BJ01-03, -BJ01-04, -BJ01-05, -BJ01-06, -BJ02-01, -BJ02-02, -BJ02-03, -BJ02-04, -BJ02-05, -BJ02-06, -BJ02-07. More detail is available on Adaptive's website at: http://adaptivebiotech.com/papers/nmed69531a.
Statistical analysis.
We determined statistical significance in values between experimental groups using either two-way ANOVA wherever appropriate (followed by post hoc analysis using the Holm–Bonferroni method), or one-way ANOVA. The distribution and variance were normal and similar in all groups. We considered any P > 0.05 as not significant. In all mouse ear-swelling measurements and FACS experiments, we give values as mean of independent replicates ± s.d. Two-way ANOVA with Holm–Bonferroni post hoc analysis was used for Figure 3b–g and Supplementary Figures 1d, 3d, 7d–f,h–i,n, 8e–g and 9b. One-way ANOVA was used for Figure 3h and Supplementary Figures 1b, 5b, 7g,i,k,m,o, 8b–d,h and 9c,d. Each of these experiments was replicated at least once (i.e., performed twice), and typically replicated twice or more. Numbers of T cell clones cannot be assumed to follow a normal distribution, so we performed comparisons in Supplementary Figure 6 by two-tailed Mann–Whitney U test.
References
Bevan, M.J. Memory T cells as an occupying force. Eur. J. Immunol. 41, 1192–1195 (2011).
Clark, R.A. et al. The vast majority of CLA+ T cells are resident in normal skin. J. Immunol. 176, 4431–4439 (2006).
Gebhardt, T., Mueller, S.N., Heath, W.R. & Carbone, F.R. Peripheral tissue surveillance and residency by memory T cells. Trends Immunol. 34, 27–32 (2013).
Kupper, T.S. Old and new: recent innovations in vaccine biology and skin T cells. J. Invest. Dermatol. 132, 829–834 (2012).
Lanzavecchia, A. & Sallusto, F. Understanding the generation and function of memory T cell subsets. Curr. Opin. Immunol. 17, 326–332 (2005).
Robins, H.S. et al. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood 114, 4099–4107 (2009).
Honda, T., Egawa, G., Grabbe, S. & Kabashima, K. Update of immune events in the murine contact hypersensitivity model: toward the understanding of allergic contact dermatitis. J. Invest. Dermatol. 133, 303–315 (2013).
Jiang, X. et al. Skin infection generates non-migratory memory CD8+ T(RM) cells providing global skin immunity. Nature 483, 227–231 (2012).
Liu, L., Fuhlbrigge, R.C., Karibian, K., Tian, T. & Kupper, T.S. Dynamic programming of CD8+ T cell trafficking after live viral immunization. Immunity 25, 511–520 (2006).
Kaech, S.M. & Wherry, E.J. Heterogeneity and cell-fate decisions in effector and memory CD8+ T cell differentiation during viral infection. Immunity 27, 393–405 (2007).
Pepper, M. & Jenkins, M.K. Origins of CD4+ effector and central memory T cells. Nat. Immunol. 12, 467–471 (2011).
Plumlee, C.R., Sheridan, B.S., Cicek, B.B. & Lefrancois, L. Environmental cues dictate the fate of individual CD8+ T cells responding to infection. Immunity 39, 347–356 (2013).
Tubo, N.J. et al. Single naive CD4+ T cells from a diverse repertoire produce different effector cell types during infection. Cell 153, 785–796 (2013).
Gerlach, C. et al. Heterogeneous differentiation patterns of individual CD8+ T cells. Science 340, 635–639 (2013).
Buchholz, V.R. et al. Disparate individual fates compose robust CD8+ T cell immunity. Science 340, 630–635 (2013).
Campbell, J.J., O'Connell, D.J. & Wurbel, M.A. Cutting edge: chemokine receptor CCR4 is necessary for antigen-driven cutaneous accumulation of CD4 T cells under physiological conditions. J. Immunol. 178, 3358–3362 (2007).
Robins, H. et al. Ultra-sensitive detection of rare T cell clones. J. Immunol. Methods 375, 14–19 (2012).
Sherwood, A.M. et al. Deep sequencing of the human TCRgamma and TCRbeta repertoires suggests that TCRbeta rearranges after alphabeta and gammadelta T cell commitment. Sci. Transl. Med. 3, 90ra61 (2011).
Popov, A., Mirkov, I. & Kataranovski, M. Inflammatory and immune mechanisms in contact hypersensitivity (CHS) in rats. Immunol. Res. 52, 127–132 (2012).
Christensen, A.D. & Haase, C. Immunological mechanisms of contact hypersensitivity in mice. APMIS 120, 1–27 (2012).
Liu, L. et al. Epidermal injury and infection during poxvirus immunization is crucial for the generation of highly protective T cell-mediated immunity. Nat. Med. 16, 224–227 (2010).
Gulati, N. et al. Molecular characterization of human skin response to diphencyprone at peak and resolution phases: therapeutic insights. J. Invest. Dermatol. 134, 2531–2540 (2014).
Watanabe, R. et al. Human skin is protected by four functionally and phenotypically discrete populations of resident and recirculating memory T cells. Sci. Transl. Med. 7, 279ra239 (2015).
Moon, J.J. et al. Quantitative impact of thymic selection on Foxp3+ and Foxp3− subsets of self-peptide/MHC class II-specific CD4+ T cells. Proc. Natl. Acad. Sci. USA 108, 14602–14607 (2011).
Mackay, L.K. et al. The developmental pathway for CD103+CD8+ tissue-resident memory T cells of skin. Nat. Immunol. 14, 1294–1301 (2013).
Acknowledgements
This work was supported by funding from the US National Institutes of Health (R01 AR065807 to T.S.K., TR01 AI097128 to T.S.K. and R.A.C., R01 AR063962 and R01 AR056720 to R.A.C.). O.G. was supported in part by the Fondation Suisse pour les Bourses en Médecine et Biologie (PASMP3-132622-1).
Author information
Authors and Affiliations
Contributions
O.G. and T.S.K. conceived and planned the study, reviewed experiments, analyzed data, and wrote the paper. O.G., R.O.E., X.J., N.G. and S.N. performed experiments. H.R., R.O.E., C.D. and J.G.K. helped plan experiments and analyze data, and R.A.C. helped analyze data and edit the paper.
Corresponding author
Ethics declarations
Competing interests
R.O.E. and C.D. have full-time employment and equity ownership at Adaptive Biotechnologies Corporation. H.R. has consultancy, patents and royalties, and equity ownership at Adaptive Biotechnologies Corporation (US patent application number: 20140256567).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–10 (PDF 1963 kb)
Rights and permissions
About this article
Cite this article
Gaide, O., Emerson, R., Jiang, X. et al. Common clonal origin of central and resident memory T cells following skin immunization. Nat Med 21, 647–653 (2015). https://doi.org/10.1038/nm.3860
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.3860