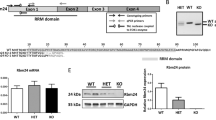
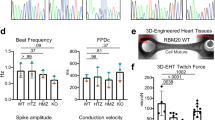
Abstract
Alternative splicing has a major role in cardiac adaptive responses, as exemplified by the isoform switch of the sarcomeric protein titin, which adjusts ventricular filling. By positional cloning using a previously characterized rat strain with altered titin mRNA splicing, we identified a loss-of-function mutation in the gene encoding RNA binding motif protein 20 (Rbm20) as the underlying cause of pathological titin isoform expression. The phenotype of Rbm20-deficient rats resembled the pathology seen in individuals with dilated cardiomyopathy caused by RBM20 mutations. Deep sequencing of the human and rat cardiac transcriptome revealed an RBM20-dependent regulation of alternative splicing. In addition to titin (TTN), we identified a set of 30 genes with conserved splicing regulation between humans and rats. This network is enriched for genes that have previously been linked to cardiomyopathy, ion homeostasis and sarcomere biology. Our studies emphasize the key role of post-transcriptional regulation in cardiac function and provide mechanistic insights into the pathogenesis of human heart failure.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Chen, M. & Manley, J.L. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol. 10, 741–754 (2009).
Lin, S. & Fu, X.-D. SR proteins and related factors in alternative splicing. Adv. Exp. Med. Biol. 623, 107–122 (2007).
Cooper, T.A., Wan, L. & Dreyfuss, G. RNA and disease. Cell 136, 777–793 (2009).
Lukong, K.E., Chang, K.-wei, Khandjian, E.W. & Richard, S. RNA-binding proteins in human genetic disease. Trends Genet. 24, 416–425 (2008).
Wang, G.S. & Cooper, T.A. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat. Rev. Genet. 8, 749–761 (2007).
Gerull, B. et al. Mutations of TTN, encoding the giant muscle filament titin, cause familial dilated cardiomyopathy. Nat. Genet. 30, 201–204 (2002).
Makarenko, I. et al. Passive stiffness changes caused by upregulation of compliant titin isoforms in human dilated cardiomyopathy hearts. Circ. Res. 95, 708–716 (2004).
Lahmers, S., Wu, Y., Call, D.R., Labeit, S. & Granzier, H. Developmental control of titin isoform expression and passive stiffness in fetal and neonatal myocardium. Circ. Res. 94, 505–513 (2004).
Opitz, C.A., Leake, M.C., Makarenko, I., Benes, V. & Linke, W.A. Developmentally regulated switching of titin size alters myofibrillar stiffness in the perinatal heart. Circ. Res. 94, 967–975 (2004).
Warren, C.M., Krzesinski, P.R., Campbell, K.S., Moss, R.L. & Greaser, M.L. Titin isoform changes in rat myocardium during development. Mech. Dev. 121, 1301–1312 (2004).
Yamasaki, R. et al. Protein kinase A phosphorylates titin's cardiac-specific N2B domain and reduces passive tension in rat cardiac myocytes. Circ. Res. 90, 1181–1188 (2002).
Cazorla, O. et al. Differential expression of cardiac titin isoforms and modulation of cellular stiffness. Circ. Res. 86, 59–67 (2000).
Wu, Y., Peng, J., Campbell, K.B., Labeit, S. & Granzier, H. Hypothyroidism leads to increased collagen-based stiffness and re-expression of large cardiac titin isoforms with high compliance. J. Mol. Cell. Cardiol. 42, 186–195 (2007).
Krüger, M. et al. Thyroid hormone regulates developmental titin isoform transitions via the phosphatidylinositol-3-kinase/AKT pathway. Circ. Res. 102, 439–447 (2008).
Krüger, M., Babicz, K., von Frieling-Salewsky, M. & Linke, W.A. Insulin signaling regulates cardiac titin properties in heart development and diabetic cardiomyopathy. J. Mol. Cell. Cardiol. 48, 910–916 (2010).
Greaser, M.L. et al. Mutation that dramatically alters rat titin isoform expression and cardiomyocyte passive tension. J. Mol. Cell. Cardiol. 44, 983–991 (2008).
Brauch, K.M. et al. Mutations in ribonucleic acid binding protein gene cause familial dilated cardiomyopathy. J. Am. Coll. Cardiol. 54, 930–941 (2009).
Li, D. et al. Identification of novel mutations in RBM20 in patients with dilated cardiomyopathy. Clin. Transl. Sci. 3, 90–97 (2010).
Greaser, M.L. et al. Developmental changes in rat cardiac titin/connectin: transitions in normal animals and in mutants with a delayed pattern of isoform transition. J. Muscle Res. Cell Motil. 26, 325–332 (2005).
Gama-Carvalho, M. et al. Targeting of U2AF65 to sites of active splicing in the nucleus. J. Cell Biol. 137, 975–987 (1997).
Okano, H.J. & Darnell, R.B. A hierarchy of Hu RNA binding proteins in developing and adult neurons. J. Neurosci. 17, 3024–3037 (1997).
Polydorides, A.D., Okano, H.J., Yang, Y.Y., Stefani, G. & Darnell, R.B. A brain-enriched polypyrimidine tract-binding protein antagonizes the ability of Nova to regulate neuron-specific alternative splicing. Proc. Natl. Acad. Sci. USA 97, 6350–6355 (2000).
Xu, X. et al. ASF/SF2-regulated CaMKIIdelta alternative splicing temporally reprograms excitation-contraction coupling in cardiac muscle. Cell 120, 59–72 (2005).
Wang, D., Papp, A.C., Binkley, P.F., Johnson, J.A. & Sadée, W. Highly variable mRNA expression and splicing of L-type voltage-dependent calcium channel α subunit 1C in human heart tissues. Pharmacogenet. Genomics 16, 735–745 (2006).
Zahler, A.M., Lane, W.S., Stolk, J.A. & Roth, M.B. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 6, 837–847 (1992).
Tacke, R. & Manley, J.L. Determinants of SR protein specificity. Curr. Opin. Cell Biol. 11, 358–362 (1999).
Lin, S., Xiao, R., Sun, P., Xu, X. & Fu, X.-D. Dephosphorylation-dependent sorting of SR splicing factors during mRNP maturation. Mol. Cell 20, 413–425 (2005).
Zhong, X.-Y., Ding, J.-H., Adams, J.A., Ghosh, G. & Fu, X.-D. Regulation of SR protein phosphorylation and alternative splicing by modulating kinetic interactions of SRPK1 with molecular chaperones. Genes Dev. 23, 482–495 (2009).
Toko, H. et al. Ca2-almodulin–dependent kinase IIΔ causes heart failure by accumulation of p53 in dilated cardiomyopathy. Circulation 122, 891–899 (2010).
Cheng, H. et al. Selective deletion of long but not short Cypher isoforms leads to late-onset dilated cardiomyopathy. Hum. Mol. Genet. 20, 1751–1762 (2011).
Tang, Z.Z. et al. Regulation of the mutually exclusive exons 8a and 8 in the CaV1.2 calcium channel transcript by polypyrimidine tract–binding protein. J. Biol. Chem. 286, 10007–10016 (2011).
Itoh-Satoh, M. et al. Titin mutations as the molecular basis for dilated cardiomyopathy. Biochem. Biophys. Res. Commun. 291, 385–393 (2002).
Neagoe, C. et al. Titin isoform switch in ischemic human heart disease. Circulation 106, 1333–1341 (2002).
Williams, L. et al. Titin isoform expression in aortic stenosis. Clin. Sci. 117, 237–242 (2009).
Chaturvedi, R.R. et al. Passive stiffness of myocardium from congenital heart disease and implications for diastole. Circulation 121, 979–988 (2010).
Ding, J.-H. et al. Dilated cardiomyopathy caused by tissue-specific ablation of SC35 in the heart. EMBO J. 23, 885–896 (2004).
Feng, Y. et al. SRp38 regulates alternative splicing and is required for Ca2+ handling in the embryonic heart. Dev. Cell 16, 528–538 (2009).
Wagner, S. et al. Ca2+/calmodulin-dependent protein kinase II regulates cardiac Na+ channels. J. Clin. Invest. 116, 3127–3138 (2006).
Vatta, M. et al. Mutations in Cypher/ZASP in patients with dilated cardiomyopathy and left ventricular non-compaction. J. Am. Coll. Cardiol. 42, 2014–2027 (2003).
Arimura, T. et al. A Cypher/ZASP mutation associated with dilated cardiomyopathy alters the binding affinity to protein kinase C. J. Biol. Chem. 279, 6746–6752 (2004).
Arimura, T. et al. Impaired binding of ZASP/Cypher with phosphoglucomutase 1 is associated with dilated cardiomyopathy. Cardiovasc. Res. 83, 80–88 (2009).
Roger, V.L. et al. Trends in heart failure incidence and survival in a community-based population. J. Am. Med. Assoc. 292, 344–350 (2004).
Mahon, N.G. et al. Echocardiographic evaluation in asymptomatic relatives of patients with dilated cardiomyopathy reveals preclinical disease. Ann. Intern. Med. 143, 108–115 (2005).
Liew, C.C. & Dzau, V.J. Molecular genetics and genomics of heart failure. Nat. Rev. Genet. 5, 811–825 (2004).
Matlin, A.J., Clark, F. & Smith, C.W.J. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell Biol. 6, 386–398 (2005).
Saar, K. et al. SNP and haplotype mapping for genetic analysis in the rat. Nat. Genet. 40, 560–566 (2008).
Hardenbol, P. et al. Multiplexed genotyping with sequence-tagged molecular inversion probes. Nat. Biotechnol. 21, 673–678 (2003).
Hardenbol, P. et al. Highly multiplexed molecular inversion probe genotyping: over 10,000 targeted SNPs genotyped in a single tube assay. Genome Res. 15, 269–275 (2005).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995).
Trapnell, C., Pachter, L. & Salzberg, S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009).
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010).
Acknowledgements
We are grateful to B. Goldbrich, O. Hummel, S. Lubitz, S. Makino, G. Patone, S. Blachut, R. Plehm, S. Probst, S. Schmidt and M. Wehle for expert technical assistance. R. Hetzer (Deutsches Herzzentrum Berlin) generously provided human cardiac tissue. Mammalian expression vectors for PTBP1 and HuD were gifts from R. Darnell, The Rockefeller University. This work was supported by the US National Institutes of Health grants HL77196 (M.L.G.) and HL075431 (C.A.M.), the Deutsche Forschungsgemeinschaft, Bonn, Germany and the European Research Council grant StG282078 (M.G.), the German Ministry of Science and Education (Nationales Genomforschungsnetz, NGFN-Plus Heart Failure Network) and EURATRANS (HEALTH-F4-2010-241504) (N.H.) and the American Heart Association (P.T.E.).
Author information
Authors and Affiliations
Contributions
M.L.G., N.H. and M.G. designed the research. W.G., S.S., M.H.R., M.L., T.G., H.M., H.S., S.L., A.M.P., V.D., P.V., S.K., B.G., L.T., V.R.-Z., T.A.H., K.W.S., G.W.D., P.T.E., C.A.M., B.S., R.F., A.P., C.O. and K.S. performed the research. W.G., S.S., M.L.G., H.S., M.H.R., M.L., T.G., H.M., S.L., A.M.P., V.D., P.V., S.K., B.G., L.T., V.R.-Z., T.A.H., K.W.S., G.W.D., P.T.E., C.A.M., B.S., R.F., A.P., C.O., K.S. and M.G. performed data analysis. W.G., T.G., H.M., H.S., S.L., A.M.P., V.D., P.V., S.K., B.G., L.T., V.R.-Z., T.A.H., K.W.S., G.W.D., P.T.E., C.A.M., B.S., R.F., A.P., C.O. and K.S. provided discussion and advice. V.R.-Z., A.P. and C.O. provided patient material. S.S., M.H.R., T.G., H.S. and M.G. performed the bioinformatics analysis. S.S., M.L.G., M.H.R., N.H. and M.G. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–11, Supplementary Tables 1–6 and Supplementary Methods (PDF 5285 kb)
Rights and permissions
About this article
Cite this article
Guo, W., Schafer, S., Greaser, M. et al. RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat Med 18, 766–773 (2012). https://doi.org/10.1038/nm.2693
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.2693