Abstract
Glutamine metabolism provides synergistic support for macrophage activation and elicitation of desirable immune responses; however, the underlying mechanisms regulated by glutamine metabolism to orchestrate macrophage activation remain unclear. Here we show that the production of α-ketoglutarate (αKG) via glutaminolysis is important for alternative (M2) activation of macrophages, including engagement of fatty acid oxidation (FAO) and Jmjd3-dependent epigenetic reprogramming of M2 genes. This M2-promoting mechanism is further modulated by a high αKG/succinate ratio, whereas a low ratio strengthens the proinflammatory phenotype in classically activated (M1) macrophages. As such, αKG contributes to endotoxin tolerance after M1 activation. This study reveals new mechanistic regulations by which glutamine metabolism tailors the immune responses of macrophages through metabolic and epigenetic reprogramming.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
References
Murray, P.J. et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity 41, 14–20 (2014).
Ivashkiv, L.B. Epigenetic regulation of macrophage polarization and function. Trends Immunol. 34, 216–223 (2013).
Biswas, S.K. & Lopez-Collazo, E. Endotoxin tolerance: new mechanisms, molecules and clinical significance. Trends Immunol. 30, 475–487 (2009).
Sawhney, S., Woo, P. & Murray, K.J. Macrophage activation syndrome: a potentially fatal complication of rheumatic disorders. Arch. Dis. Child. 85, 421–426 (2001).
Ho, P.C., Tsui, Y.C., Feng, X., Greaves, D.R. & Wei, L.N. NF-κB-mediated degradation of the coactivator RIP140 regulates inflammatory responses and contributes to endotoxin tolerance. Nat. Immunol. 13, 379–386 (2012).
O'Neill, L.A. & Pearce, E.J. Immunometabolism governs dendritic cell and macrophage function. J. Exp. Med. 213, 15–23 (2016).
O'Neill, L.A., Kishton, R.J. & Rathmell, J. A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 16, 553–565 (2016).
Tannahill, G.M. et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature 496, 238–242 (2013).
Liu, L. et al. Proinflammatory signal suppresses proliferation and shifts macrophage metabolism from Myc-dependent to HIF1α-dependent. Proc. Natl. Acad. Sci. USA 113, 1564–1569 (2016).
Mills, E.L. et al. Succinate Dehydrogenase Supports Metabolic Repurposing of Mitochondria to Drive Inflammatory Macrophages. Cell 167, 457–470.e413 (2016).
Huang, S.C. et al. Cell-intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nat. Immunol. 15, 846–855 (2014).
Vats, D. et al. Oxidative metabolism and PGC-1beta attenuate macrophage-mediated inflammation. Cell Metab. 4, 13–24 (2006).
Covarrubias, A.J. et al. Akt-mTORC1 signaling regulates Acly to integrate metabolic input to control of macrophage activation. eLife 5, e11612 (2016).
Huang, S.C. et al. Metabolic Reprogramming Mediated by the mTORC2-IRF4 Signaling Axis Is Essential for Macrophage Alternative Activation. Immunity 45, 817–830 (2016).
Jha, A.K. et al. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 42, 419–430 (2015).
Fendt, S.M. et al. Reductive glutamine metabolism is a function of the α-ketoglutarate to citrate ratio in cells. Nat. Commun. 4, 2236 (2013).
Carey, B.W., Finley, L.W., Cross, J.R., Allis, C.D. & Thompson, C.B. Intracellular α-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature 518, 413–416 (2015).
Bricker, D.K. et al. A mitochondrial pyruvate carrier required for pyruvate uptake in yeast, Drosophila, and humans. Science 337, 96–100 (2012).
Ishii, M. et al. Epigenetic regulation of the alternatively activated macrophage phenotype. Blood 114, 3244–3254 (2009).
Satoh, T. et al. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat. Immunol. 11, 936–944 (2010).
Kruidenier, L. et al. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature 488, 404–408 (2012).
Platt, R.J. et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455 (2014).
TeSlaa, T. α-Ketoglutarate Accelerates the Initial Differentiation of Primed Human Pluripotent Stem Cells. Cell Metab. 24, 485–493 (2016).
Fraisl, P., Aragonés, J. & Carmeliet, P. Inhibition of oxygen sensors as a therapeutic strategy for ischaemic and inflammatory disease. Nat. Rev. Drug Discov. 8, 139–152 (2009).
Cummins, E.P. et al. Prolyl hydroxylase-1 negatively regulates IkappaB kinase-beta, giving insight into hypoxia-induced NFkappaB activity. Proc. Natl. Acad. Sci. USA 103, 18154–18159 (2006).
Takeda, Y. et al. Macrophage skewing by Phd2 haplodeficiency prevents ischaemia by inducing arteriogenesis. Nature 479, 122–126 (2011).
Rodríguez-Prados, J.C. et al. Substrate fate in activated macrophages: a comparison between innate, classic, and alternative activation. J. Immunol. 185, 605–614 (2010).
Shalova, I.N. et al. Human monocytes undergo functional re-programming during sepsis mediated by hypoxia-inducible factor-1α. Immunity 42, 484–498 (2015).
Su, X. et al. Interferon-γ regulates cellular metabolism and mRNA translation to potentiate macrophage activation. Nat. Immunol. 16, 838–849 (2015).
Cheng, S.C. et al. Broad defects in the energy metabolism of leukocytes underlie immunoparalysis in sepsis. Nat. Immunol. 17, 406–413 (2016).
Chen, J. & Ivashkiv, L.B. IFN-γ abrogates endotoxin tolerance by facilitating Toll-like receptor-induced chromatin remodeling. Proc. Natl. Acad. Sci. USA 107, 19438–19443 (2010).
De Santa, F. et al. Jmjd3 contributes to the control of gene expression in LPS-activated macrophages. EMBO J. 28, 3341–3352 (2009).
Pan, D. et al. Jmjd3-Mediated H3K27me3 Dynamics Orchestrate Brown Fat Development and Regulate White Fat Plasticity. Dev. Cell 35, 568–583 (2015).
Zhang, Q. et al. Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6. Nature 525, 389–393 (2015).
Saeed, S. et al. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science 345, 1251086 (2014).
Arts, R.J. et al. Glutaminolysis and Fumarate Accumulation Integrate Immunometabolic and Epigenetic Programs in Trained Immunity. Cell Metab. 24, 807–819 (2016).
del Fresno, C. et al. Tumor cells deactivate human monocytes by up-regulating IL-1 receptor associated kinase-M expression via CD44 and TLR4. J. Immunol. 174, 3032–3040 (2005).
Hagemann, T. et al. “Re-educating” tumor-associated macrophages by targeting NF-kB. J. Exp. Med. 205, 1261–1268 (2008).
Mantovani, A. & Sica, A. Macrophages, innate immunity and cancer: balance, tolerance, and diversity. Curr. Opin. Immunol. 22, 231–237 (2010).
Wise, D.R. & Thompson, C.B. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem. Sci. 35, 427–433 (2010).
Altman, B.J., Stine, Z.E. & Dang, C.V. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat. Rev. Cancer 16, 619–634 (2016).
Ye, D., Ma, S., Xiong, Y. & Guan, K.L. R-2-hydroxyglutarate as the key effector of IDH mutations promoting oncogenesis. Cancer Cell 23, 274–276 (2013).
Black, J.C., Van Rechem, C. & Whetstine, J.R. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol. Cell 48, 491–507 (2012).
Clever, D. et al. Oxygen sensing by T cells establishes an immunologically tolerant metastatic niche. Cell 166, 1117–1131.e14 (2016).
Christen, S. et al. Breast cancer-derived lung metastases show increased pyruvate carboxylase-dependent anaplerosis. Cell Rep. 17, 837–848 (2016).
Acknowledgements
We thank F. Cottard and C.-P. Lin for technical help and P. Romero and C. Hess for discussion. Supported by Swiss National Science Foundation project grant (31003A_163204), the Swiss Institute for Experimental Cancer Research (26075483), the Harry J. Lloyd Charitable Foundation, the Swiss Cancer Foundation (KFS-3949-08-2016) and a Melanoma Research Alliance Young Investigator Award to P.-C.H. S.-M.F. is supported by a Flanders Research Foundation (FWO) research grant and by Eugène Yourassowsky Schenking. J.I. is supported by the University of Lausanne. H.-D.H. is supported by the Ministry of Science and Technology of Taiwan (MOST105-2627-M-009-007 and MOST103-2628-B-009-001-MY3). T.C. is supported by a University of Lausanne FBM PhD fellowship. M.V. is supported by the National scholarship program of the Slovak Republic.
Author information
Authors and Affiliations
Contributions
P.-S.L., H.W., X.L., T.C., T.T., S.C., G.D.C.,W.-C.C., M.V., C.M., K.D. and J.I. performed experiments. P.-S.L., H.W., S.C., C.-H.C., M.M., H.-D.H., S.-M.F., J.I. and P.-C.H. analyzed results. P.-S.L., H.W. and P.-C.H. designed the studies. P.-S.L. and P.-C.H. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
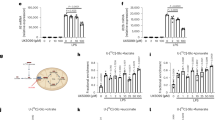
Supplementary Figure 1 αKG produced from glutaminolysis supports M2 phenotype but inhibits M1 activation.
(a) An illustration of α-ketoglutarate metabolic pathway controlled by genes listed in (Fig. 1d). (b, c) Quantitative PCR (qPCR) analysis of relative mRNA expression of M2 marker genes in BMDMs stimulated with IL-4 (b) and M1 marker genes in BMDMs stimulated with LPS (c) in glutamine-replete media supplemented with indicated treatments for 6h. Compound 968 (10μM); Dimethyl-α-ketoglutarate (DM-αKG; 1mM). (d, e) qPCR analysis of the relative mRNA expression of M2 markers genes (d) and arginase activity (e) in BMDMs stimulated with or without IL-4 in glutamine-depleted media plus escalating doses of DM-αKG for 6h. (f, g) qPCR analysis of the relative mRNA expression of M1 markers genes (f) and ELISA of IL-1β and TNFα cytokine production (g) in BMDMs stimulated with or without LPS in glutamine-depleted media plus escalating doses of DM-αKG for 6h. (h) The effects of glutaminase 1 inhibitor (BPTES; 10mM) and supplementation of DM-aKG (1mM) on IL-1β secretion in LPS stimulated macrophages in the presence of Nigericin (left panel) or MSU (monosodium urate crystal) (right panel) were analyzed by ELISA. *P < 0.05 is determine by unpaired, two-tailed Student's t-test. Data shown are representative from three independent experiments (b, c, d, e, f; mean ± s.d.), from two independent experiments (g; mean ± s.d.) and cumulative results of two independent experiments (h; mean ± s.d.).
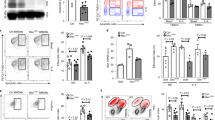
Supplementary Figure 2 αKG does not modulate IL-4-induced STAT6 activation.
(a) Immunoblot analysis of phosphor-STAT6 and STAT6 in BMDMs stimulated with IL-4 in glutamine-replete (+Gln.) or glutamine-depleted (-Gln) media for 0-2h. β-actin served as loading control. (b) Immunoblot analysis of phosphor-STAT6 and STAT6 in BMDMs stimulated with IL-4 in the presence of absence of BPTES in glutamine-replete media for 0-2h. (c) Immunoblot analysis of phosphor-STAT6 and STAT6 in BMDMs activated with IL-4 in in glutamine-depleted media supplemented with control vehicle (Ctrl) or DM-αKG (1mM). (d, e) qPCR analysis of relative mRNA expression of M2 marker genes (d) and arginase activity (e) in BMDMs stimulated with IL-4 in the indicated cultures for 6h. (f) Immunoblot analysis of Jmjd3 and β-actin 6 in BMDMs derived from Cas9-LysM-Cre mice transduced with lentivirus harboring control sgRNAs or Jmjd3-targeting sgRNAs. Data shown are representative from three independent experiments (a, b, c, d, e) and from two independent experiments (f).
Supplementary Figure 3 αKG/succinate ratio generated by macrophage activation regulates macrophage immune responses.
(a) The expression of intracellular succinate (Suc.) and α-ketoglutarate (αKG) in BMDMs with IL-4 or LPS for 18h, followed by metabolite extraction mentioned in methods and then measured with mass spectrometry. (b) Arginase activity in BMDMs stimulated with IL-4 in glutamine-depleted media supplemented with diethyl-succinate (DE-Suc.; 5mM) plus indicated concentration of DM-αKG for 6h. (c) ELISA of IL-1β in BMDMs stimulated with LPS in glutamine-depleted media supplemented with diethyl-succinate (DE-Suc.; 5mM) plus indicated concentration of DM-αKG for 6h. (d) Arginase activity in BMDMs stimulated with IL-4 in glutamine-depleted media supplemented with DM-αKG plus indicated concentration of DE-Suc. for 6h. (e) ELISA of IL-1β in BMDMs stimulated with LPS in glutamine-depleted media supplemented with DM-αKG plus indicated concentration of DE-Suc. for 6h. *P < 0.05 is determine by unpaired, two-tailed Student's t-test. Data shown are representative from three independent experiments (a, b, d; mean ± s.d.) and from two independent experiments (c, e; mean ± s.d.).
Supplementary Figure 4 Glutaminolysis suppresses NF-κB nuclear translocation upon LPS treatment.
(a) Immunoblot analysis of nuclear NF-κB p65 in BMDMs stimulated with LPS plus control vehicle (Ctrl) or BPTES in glutamine-replete media for 0-60 mins, followed by nuclear fraction isolation. Lamin A/C served as loading control for nuclear fraction lysate and β-actin served as loading control for whole cell lysate (WCL). (b) Immunoblot analysis of IKKβ in BMDMs left untransduced (Ctrl) or transduced with lentivirus harboring green fluorescence protein (GFP)-fused wild type (Wt) or P191A mutant form (P191A) of IKKβ. β-actin served as loading control. Data shown are representative from three independent experiments (a) and from two independent experiments (b).
Supplementary Figure 5 Glutaminolysis during LPS priming is essential for induction of endotoxin tolerance via impairment of the NF-κB pathway.
(a) A schematic diagram of in vitro endotoxin tolerance experimental design. (b) qPCR analysis of relative mRNA expression in BMDMs stimulated with or without LPS (10ng/ml) in glutamine-replete media supplemented with control vehicle (Ctrl) or BPTES for 18h and then re-challenged with LPS (10ng/ml) for another 6h. (c) A schematic diagram of in vitro endotoxin tolerance experimental design. BMDMs were stimulated with or without LPS (10ng/ml) in glutamine-depleted (Gln.-depleted) media supplemented with or without DM-αKG for 18h and then re-challenged with LPS (10ng/ml). (d, e) IL-1β (d) and TNFα (e) production in BMDMs stimulated with LPS as described in (c), followed by ELISA analysis. (f) A heat map showing expression of genes encoding tissue repairing and antimicrobial molecules in BMDMs stimulated with LPS for 18h in glutamine-replete media (ET w/ Gln.), glutamine-depleted media (ET w/o Gln.) or glutamine-depleted media supplemented with 1mM DM-αKG (ET w/o Gln.+ DM-αKG), assessed by RNA-sequencing. (g) A heat map showing metabolome changes associated with induction of endotoxin tolerance, based on metabolite expression between non-tolerant macrophage (LPS w/ Gln.; LPS stimulation for 6h in glutamine-replete media) and tolerant macrophages (ET w/ Gln.; endotoxin tolerant macrophages stimulated with LPS for 6h in glutamine-replete media). The metabolites of this list in BMDMs stimulated with LPS in glutamine-depleted media (ET w/o Gln.) or glutamine-depleted media supplemented with DM-αKG (ET w/o Gln.+ DM-αKG) for 18h and then re-activated with LPS for another 6h were also included in this heat map. (h) The expression of lactate and pyruvate in BMDMs of indicated groups were determined by mass spectrometry. *P < 0.05 is determine by unpaired, two-tailed Student's t-test. Data shown are representative from three independent experiments (b; mean ± s.d.), cumulative results from two independent experiments (d, e, f; mean ± s.d.) and combined from three independent experiments and normalize to LPS w/ Gln. group (g, h; mean ± s.d.).
Supplementary Figure 6 Glutamine metabolism during LPS priming prevents reactivation of proinflammatory genes in a Jmjd3-independent manner.
(a, b, c) Immunoblots of proximal signaling pathways of toll-like receptor 4 in BMDMs treated as described in figure 6a (a) or treated in glutamine-replete media supplemented with control vehicle (Ctrl) or BPTES (b), or treated in glutamine-depleted media supplemented with or without DM-αKG (c), for 18h and then re-stimulated with second exposure of LPS for 0-60 mins. β-actin served as loading control. (d) Immunoblots of NF-κB p65 in BMDMs treated with LPS in glutamine-replete media supplemented with or without BPTES for 18h, followed by re-stimulated with second exposure of LPS for 0-60 mins. (e) Immunoblots of NF-κB p65 in BMDMs treated with first exposure of LPS in glutamine-replete (+Gln.) or glutamine-depleted (-Gln.) media plus control vehicle or DM-αKG for 18h. After washing, BMDMs were then re-stimulated with second exposure of LPS for indicated period. Those BMDMs did not receive first LPS treatment represent control groups for intact responsiveness to LPS-induced nuclear translocation of NF-κB. (f) qPCR analysis of relative mRNA expression of M1 marker genes in Cas9-expressing BMDMs transduced with lentivirus harboring control sgRNAs or Jmjd3-targeting sgRNAs. These BMDMs were treated with or without LPS in the indicated conditions for 18h and then re-challenged with LPS re-exposure in glutamine-replete media for another 6h. (g) A schematic diagram of in vivo endotoxin tolerance assay. *P < 0.05 is determine by unpaired, two-tailed Student's t-test. Data shown are representative from three independent experiments (a, b, c, d, e, f; mean ± s.d.).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 and Supplementary Tables 1–3 (PDF 1390 kb)
Rights and permissions
About this article
Cite this article
Liu, PS., Wang, H., Li, X. et al. α-ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming. Nat Immunol 18, 985–994 (2017). https://doi.org/10.1038/ni.3796
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.3796