Abstract
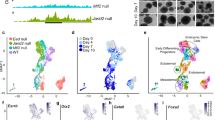
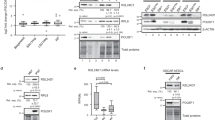
The molecular controls that govern the differentiation of embryonic stem (ES) cells remain poorly understood. DGCR8 is an RNA-binding protein that assists the RNase III enzyme Drosha in the processing of microRNAs (miRNAs), a subclass of small RNAs. Here we study the role of miRNAs in ES cell differentiation by generating a Dgcr8 knockout model. Analysis of mouse knockout ES cells shows that DGCR8 is essential for biogenesis of miRNAs. On the induction of differentiation, DGCR8-deficient ES cells do not fully downregulate pluripotency markers and retain the ability to produce ES cell colonies; however, they do express some markers of differentiation. This phenotype differs from that reported for Dicer1 knockout cells, suggesting that Dicer has miRNA-independent roles in ES cell function. Our findings indicate that miRNAs function in the silencing of ES cell self-renewal that normally occurs with the induction of differentiation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Valencia-Sanchez, M.A., Liu, J., Hannon, G.J. & Parker, R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 20, 515–524 (2006).
Lewis, B.P., Burge, C.B. & Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20 (2005).
Wienholds, E. & Plasterk, R.H. MicroRNA function in animal development. FEBS Lett. 579, 5911–5922 (2005).
Kanellopoulou, C. et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 19, 489–501 (2005).
Bernstein, E. et al. Dicer is essential for mouse development. Nat. Genet. 35, 215–217 (2003).
Hammond, S.M. Dicing and slicing: the core machinery of the RNA interference pathway. FEBS Lett. 579, 5822–5829 (2005).
Hutvagner, G. et al. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293, 834–838 (2001).
Tomari, Y. & Zamore, P.D. MicroRNA biogenesis: Drosha can't cut it without a partner. Curr. Biol. 15, R61–R64 (2005).
Denli, A.M., Tops, B.B., Plasterk, R.H., Ketting, R.F. & Hannon, G.J. Processing of primary microRNAs by the Microprocessor complex. Nature 432, 231–235 (2004).
Gregory, R.I. et al. The Microprocessor complex mediates the genesis of microRNAs. Nature 432, 235–240 (2004).
Han, J. et al. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 18, 3016–3027 (2004).
Landthaler, M., Yalcin, A. & Tuschl, T. The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr. Biol. 14, 2162–2167 (2004).
Han, J. et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 125, 887–901 (2006).
Yi, R., Qin, Y., Macara, I.G. & Cullen, B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 17, 3011–3016 (2003).
Lund, E., Guttinger, S., Calado, A., Dahlberg, J.E. & Kutay, U. Nuclear export of microRNA precursors. Science 303, 95–98 (2004).
Wu, H., Xu, H., Miraglia, L.J. & Crooke, S.T. Human RNase III is a 160-kDa protein involved in preribosomal RNA processing. J. Biol. Chem. 275, 36957–36965 (2000).
Houbaviy, H.B., Murray, M.F. & Sharp, P.A. Embryonic stem cell-specific microRNAs. Dev. Cell 5, 351–358 (2003).
Houbaviy, H.B., Dennis, L., Jaenisch, R. & Sharp, P.A. Characterization of a highly variable eutherian microRNA gene. RNA 11, 1245–1257 (2005).
Hatfield, S.D. et al. Stem cell division is regulated by the microRNA pathway. Nature 435, 974–978 (2005).
Hebert, J.M., Boyle, M. & Martin, G.R. mRNA localization studies suggest that murine FGF-5 plays a role in gastrulation. Development 112, 407–415 (1991).
Rathjen, J. et al. Formation of a primitive ectoderm like cell population, EPL cells, from ES cells in response to biologically derived factors. J. Cell Sci. 112, 601–612 (1999).
Pelton, T.A., Sharma, S., Schulz, T.C., Rathjen, J. & Rathjen, P.D. Transient pluripotent cell populations during primitive ectoderm formation: correlation of in vivo and in vitro pluripotent cell development. J. Cell Sci. 115, 329–339 (2002).
Kaji, K. et al. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat. Cell Biol. 8, 285–292 (2006).
Feldman, N. et al. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat. Cell Biol. 8, 188–194 (2006).
Murchison, E.P., Partridge, J.F., Tam, O.H., Cheloufi, S. & Hannon, G.J. Characterization of Dicer-deficient murine embryonic stem cells. Proc. Natl. Acad. Sci. USA 102, 12135–12140 (2005).
Blelloch, R.H. et al. Nuclear cloning of embryonal carcinoma cells. Proc. Natl. Acad. Sci. USA 101, 13985–13990 (2004).
Lau, N.C., Lim, L.P., Weinstein, E.G. & Bartel, D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294, 858–862 (2001).
Di Cristofano, A., Pesce, B., Cordon-Cardo, C. & Pandolfi, P.P. Pten is essential for embryonic development and tumor suppression. Nat. Genet. 19, 348–355 (1998).
Acknowledgements
We thank C. Beard and the Anita Sil Laboratory for technical advice; J. Dausman for technical help; J. Reiter and N. Shomron for reagents and M. Ramalho-Santos, J. Reiter and members of the Blelloch Laboratory for critically reading the manuscript. The primers for Oct4, Rex1, Gapdh, T (brachyury) and Krt18 were gifts from J. Reiter (University of California San Francisco) This work was supported by grants to R.B. from the University of California San Francisco Urology Department, the Sandler Foundation, the Lance Armstrong Foundation and the National Institutes of Health (K08 NS48118). Y.W. is supported by the California Institute of Regenerative Medicine postdoctoral fellowship program. C.M. is supported by the National Science Foundation graduate research fellowship program.
Author information
Authors and Affiliations
Contributions
Y.W. did the experiments described in Figs. 2 and 3 and quantitative PCR. C.M. did the experiments described in Fig. 5b–d. R.M. together with R.B. performed other experiments, including construction of knockout ES cells. R.J. provided reagent and infrastructure support for targeting experiments. R.B. was involved in designing the experiments and wrote the paper with help from Y.W. and C.M.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
RNA blot and microarray analysis of miRNAs. (PDF 138 kb)
Supplementary Fig. 2
Analysis of apoptosis in wild-type, DGCR8 knockout and DGCR8 rescued ES cells. (PDF 60 kb)
Supplementary Fig. 3
Quantitative real-time PCR analysis of Nanog, Afp, Vasa, Eomes and Sox1 after EB differentiation at 0, 8 and 16 d. (PDF 36 kb)
Supplementary Fig. 4
Semiquantitative RT-PCR analysis of wild-type and rescued ES cells. (PDF 48 kb)
Supplementary Fig. 5
Colony differentiation assay. (PDF 204 kb)
Supplementary Table 1
Ratio of miRNA expression for δ/flox and δ/δ relative to wild-type. (PDF 24 kb)
Supplementary Table 2
Ratio of abnormal implants from DGCR8 wt/δ × wt/δ crosses at different time points in embryonic development. (PDF 16 kb)
Supplementary Table 3
Sequences of probes used in RNA blot analysis and primers used in quantitative real-time PCR. (PDF 16 kb)
Rights and permissions
About this article
Cite this article
Wang, Y., Medvid, R., Melton, C. et al. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet 39, 380–385 (2007). https://doi.org/10.1038/ng1969
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1969