Abstract
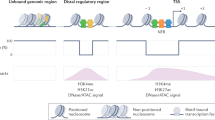
Organisms respond to changes in their environment, and many such responses are initiated at the level of gene transcription. Here, we provide evidence for a previously undiscovered mechanism for directing transcriptional regulators to new binding targets in response to an environmental change. We show that repressor-activator protein 1 (Rap1), a master regulator of yeast metabolism, binds to an expanded target set after glucose depletion despite decreasing protein levels and no evidence of posttranslational modification. Computational analysis predicts that proteins capable of recruiting the chromatin regulator Tup1 act to restrict the binding distribution of Rap1 in the presence of glucose. Deletion of the gene(s) encoding Tup1, recruiters of Tup1 or chromatin regulators recruited by Tup1 cause Rap1 to bind specifically and inappropriately to low-glucose targets. These data, combined with whole-genome measurements of nucleosome occupancy and Tup1 distribution, provide evidence for a mechanism of dynamic target specification that coordinates the genome-wide distribution of intermediate-affinity DNA sequence motifs with chromatin-mediated regulation of accessibility to those sites.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Jacob, F. & Monod, J. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 3, 318–356 (1961).
Harbison, C.T. et al. Transcriptional regulatory code of a eukaryotic genome. Nature 431, 99–104 (2004).
O'Neill, E.M., Kaffman, A., Jolly, E.R. & O'Shea, E.K. Regulation of PHO4 nuclear localization by the PHO80–PHO85 cyclin-CDK complex. Science 271, 209–212 (1996).
Zubay, G., Schwartz, D. & Beckwith, J. Mechanism of activation of catabolite-sensitive genes: a positive control system. Proc. Natl. Acad. Sci. USA 66, 104–110 (1970).
Zeitlinger, J. et al. Program-specific distribution of a transcription factor dependent on partner transcription factor and MAPK signaling. Cell 113, 395–404 (2003).
Gu, W. & Roeder, R.G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90, 595–606 (1997).
Shore, D. RAP1: a protean regulator in yeast. Trends Genet. 10, 408–412 (1994).
Lieb, J.D., Liu, X., Botstein, D. & Brown, P.O. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat. Genet. 28, 327–334 (2001).
Zhu, H. et al. Analysis of yeast protein kinases using protein chips. Nat. Genet. 26, 283–289 (2000).
Smith, R.L. & Johnson, A.D. Turning genes off by Ssn6-Tup1: a conserved system of transcriptional repression in eukaryotes. Trends Biochem. Sci. 25, 325–330 (2000).
Regnacq, M., Alimardani, P., El Moudni, B. & Berges, T. SUT1p interaction with Cyc8p(Ssn6p) relieves hypoxic genes from Cyc8p-Tup1p repression in Saccharomyces cerevisiae. Mol. Microbiol. 40, 1085–1096 (2001).
Tzamarias, D. & Struhl, K. Functional dissection of the yeast Cyc8-Tup1 transcriptional co-repressor complex. Nature 369, 758–761 (1994).
Proft, M. & Serrano, R. Repressors and upstream repressing sequences of the stress-regulated ENA1 gene in Saccharomyces cerevisiae: bZIP protein Sko1p confers HOG-dependent osmotic regulation. Mol. Cell. Biol. 19, 537–546 (1999).
De Vit, M.J., Waddle, J.A. & Johnston, M. Regulated nuclear translocation of the Mig1 glucose repressor. Mol. Biol. Cell 8, 1603–1618 (1997).
Park, S.H., Koh, S.S., Chun, J.H., Hwang, H.J. & Kang, H.S. Nrg1 is a transcriptional repressor for glucose repression of STA1 gene expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 19, 2044–2050 (1999).
Proft, M. & Struhl, K. Hog1 kinase converts the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol. Cell 9, 1307–1317 (2002).
Davie, J.K., Edmondson, D.G., Coco, C.B. & Dent, S.Y. Tup1-Ssn6 interacts with multiple class I histone deacetylases in vivo. J. Biol. Chem. 278, 50158–50162 (2003).
Watson, A.D. et al. Ssn6-Tup1 interacts with class I histone deacetylases required for repression. Genes Dev. 14, 2737–2744 (2000).
Wu, J., Suka, N., Carlson, M. & Grunstein, M. TUP1 utilizes histone H3/H2B-specific HDA1 deacetylase to repress gene activity in yeast. Mol. Cell 7, 117–126 (2001).
Green, S.R. & Johnson, A.D. Promoter-dependent roles for the Srb10 cyclin-dependent kinase and the Hda1 deacetylase in Tup1-mediated repression in Saccharomyces cerevisiae. Mol. Biol. Cell 15, 4191–4202 (2004).
Zhang, Z. & Reese, J.C. Ssn6-Tup1 requires the ISW2 complex to position nucleosomes in Saccharomyces cerevisiae. EMBO J. 23, 2246–2257 (2004).
Fazzio, T.G. et al. Widespread collaboration of Isw2 and Sin3-Rpd3 chromatin remodeling complexes in transcriptional repression. Mol. Cell. Biol. 21, 6450–6460 (2001).
Mukherjee, S. et al. Rapid analysis of the DNA-binding specificities of transcription factors with DNA microarrays. Nat. Genet. 36, 1331–1339 (2004).
Rao, B., Shibata, Y., Strahl, B.D. & Lieb, J.D. Dimethylation of histone H3 at lysine 36 demarcates regulatory and nonregulatory chromatin genome-wide. Mol. Cell. Biol. 25, 9447–9459 (2005).
Buck, M., Nobel, A. & Lieb, J. ChIPOTle: a user-friendly tool for the analysis of ChIP-chip data. Genome Biol. 6, R97 (2005).
Lee, C.K., Shibata, Y., Rao, B., Strahl, B.D. & Lieb, J.D. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 36, 900–905 (2004).
Liu, X.S., Brutlag, D.L. & Liu, J.S. An algorithm for finding protein-DNA binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nat. Biotechnol. 20, 835–839 (2002).
Proft, M., Gibbons, F.D., Copeland, M., Roth, F.P. & Struhl, K. Genomewide identification of Sko1 target promoters reveals a regulatory network that operates in response to osmotic stress in Saccharomyces cerevisiae. Eukaryot. Cell 4, 1343–1352 (2005).
Crooks, G.E., Hon, G., Chandonia, J.M. & Brenner, S.E. WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 (2004).
Haverty, P.M., Hansen, U. & Weng, Z. Computational inference of transcriptional regulatory networks from expression profiling and transcription factor binding site identification. Nucleic Acids Res. 32, 179–188 (2004).
Acknowledgements
We thank A.D. Johnson (Department of Microbiology and Immunology, University of California, San Francisco) for providing a strain. This work was supported by US National Institutes of Health grant HG002989 to M.J.B. and GM072518 to J.D.L.
Author information
Authors and Affiliations
Contributions
This study was designed by M.J.B. and J.D.L. M.J.B conducted the experiments and data analysis. M.J.B. and J.D.L. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Rap1 protein and mRNA levels decrease as glucose is depleted. (PDF 462 kb)
Supplementary Fig. 2
Confirmation of ChIP-chip results by PCR. (PDF 1488 kb)
Supplementary Fig. 3
Confirmation of ChIP-chip results at the SGA1 locus. (PDF 924 kb)
Supplementary Fig. 4
Computational screens predict the involvement of Tup1-Ssn6 proteins in blocking Rap1 binding. (PDF 290 kb)
Supplementary Fig. 5
Rap1 binding and glycolysis. (PDF 372 kb)
Supplementary Table 1
ChIP-chip experiments. (PDF 53 kb)
Supplementary Table 2
Rap1 binding targets. (XLS 71 kb)
Rights and permissions
About this article
Cite this article
Buck, M., Lieb, J. A chromatin-mediated mechanism for specification of conditional transcription factor targets. Nat Genet 38, 1446–1451 (2006). https://doi.org/10.1038/ng1917
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1917