Abstract
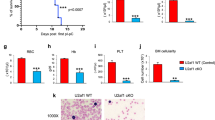
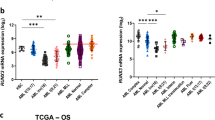
Knockdown of the transcription factor PU.1 (encoded by Sfpi1) leads to acute myeloid leukemia (AML) in mice. We examined the transcriptome of preleukemic hematopoietic stem cells (HSCs) in which PU.1 was knocked down (referred to as 'PU.1-knockdown HSCs') to identify transcriptional changes preceding malignant transformation. Transcription factors c-Jun and JunB were among the top-downregulated targets. Restoration of c-Jun expression in preleukemic cells rescued the PU.1 knockdown–initiated myelomonocytic differentiation block. Lentiviral restoration of JunB at the leukemic stage led to loss of leukemic self-renewal capacity and prevented leukemia in NOD-SCID mice into which leukemic PU.1-knockdown cells were transplanted. Examination of human individuals with AML confirmed the correlation between PU.1 and JunB downregulation. These results delineate a transcriptional pattern that precedes leukemic transformation in PU.1-knockdown HSCs and demonstrate that decreased levels of c-Jun and JunB contribute to the development of PU.1 knockdown–induced AML by blocking differentiation and increasing self-renewal. Therefore, examination of disturbed gene expression in HSCs can identify genes whose dysregulation is essential for leukemic stem cell function and that are targets for therapeutic interventions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
References
McKercher, S.R. et al. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 15, 5647–5658 (1996).
Mueller, B.U. et al. Heterozygous PU.1 mutations are associated with acute myeloid leukemia. Blood 100, 998–1007 (2002).
Vangala, R.K. et al. The myeloid master regulator transcription factor PU.1 is inactivated by AML1-ETO in t(8;21) myeloid leukemia. Blood 101, 270–277 (2003).
Gilliland, D.G. & Tallman, M.S. Focus on acute leukemias. Cancer Cell 1, 417–420 (2002).
Li, Y. et al. Regulation of the PU.1 gene by distal elements. Blood 98, 2958–2965 (2001).
Okuno, Y. et al. Potential autoregulation of transcription factor PU.1 by an upstream regulatory element. Mol. Cell. Biol. 25, 2832–2845 (2005).
Rosenbauer, F. et al. Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nat. Genet. 36, 624–630 (2004).
Rosenbauer, F., Koschmieder, S., Steidl, U. & Tenen, D.G. Effect of transcription-factor concentrations on leukemic stem cells. Blood 106, 1519–1524 (2005).
Tenen, D.G. Disruption of differentiation in human cancer: AML shows the way. Nat. Rev. Cancer 3, 89–101 (2003).
Passegue, E., Wagner, E.F. & Weissman, I.L. JunB deficiency leads to a myeloproliferative disorder arising from hematopoietic stem cells. Cell 119, 431–443 (2004).
Behre, G. et al. c-Jun is a JNK-independent coactivator of the PU.1 transcription factor. J. Biol. Chem. 274, 4939–4946 (1999).
Phinney, D.G., Tseng, S.W., Hall, B. & Ryder, K. Chromosomal integration dependent induction of junB by growth factors requires multiple flanking evolutionarily conserved sequences. Oncogene 13, 1875–1883 (1996).
Higuchi, M. et al. Expression of a conditional AML1-ETO oncogene bypasses embryonic lethality and establishes a murine model of human t(8;21) acute myeloid leukemia. Cancer Cell 1, 63–74 (2002).
Huntly, B.J. et al. MOZ-TIF2, but not BCR-ABL, confers properties of leukemic stem cells to committed murine hematopoietic progenitors. Cancer Cell 6, 587–596 (2004).
Tenen, D.G., Hromas, R., Licht, J.D. & Zhang, D.E. Transcription factors, normal myeloid development, and leukemia. Blood 90, 489–519 (1997).
Borras, F.E., Lloberas, J., Maki, R.A. & Celada, A. Repression of I-A beta gene expression by the transcription factor PU.1. J. Biol. Chem. 270, 24385–24391 (1995).
Bellon, T., Perrotti, D. & Calabretta, B. Granulocytic differentiation of normal hematopoietic precursor cells induced by transcription factor PU.1 correlates with negative regulation of the c-myb promoter. Blood 90, 1828–1839 (1997).
Wang, Q., Salman, H., Danilenko, M. & Studzinski, G.P. Cooperation between antioxidants and 1,25-dihydroxyvitamin D3 in induction of leukemia HL60 cell differentiation through the JNK/AP-1/Egr-1 pathway. J. Cell. Physiol. 204, 964–974 (2005).
Bhardwaj, G. et al. Sonic hedgehog induces the proliferation of primitive human hematopoietic cells via BMP regulation. Nat. Immunol. 2, 172–180 (2001).
Antonchuk, J., Sauvageau, G. & Humphries, R.K. HOXB4-induced expansion of adult hematopoietic stem cells ex vivo. Cell 109, 39–45 (2002).
Reya, T. et al. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature 423, 409–414 (2003).
Iwama, A. et al. Enhanced self-renewal of hematopoietic stem cells mediated by the polycomb gene product Bmi-1. Immunity 21, 843–851 (2004).
Duncan, A.W. et al. Integration of Notch and Wnt signaling in hematopoietic stem cell maintenance. Nat. Immunol. 6, 314–322 (2005).
Valk, P.J. et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N. Engl. J. Med. 350, 1617–1628 (2004).
Bullinger, L. et al. Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N. Engl. J. Med. 350, 1605–1616 (2004).
Bjerregaard, M.D., Jurlander, J., Klausen, P., Borregaard, N. & Cowland, J.B. The in vivo profile of transcription factors during neutrophil differentiation in human bone marrow. Blood 101, 4322–4332 (2003).
Rosenbauer, F. et al. Lymphoid cell growth and transformation are suppressed by a key regulatory element of the gene encoding PU.1. Nat. Genet. 38, 27–37 (2006).
Reya, T., Morrison, S.J., Clarke, M.F. & Weissman, I.L. Stem cells, cancer, and cancer stem cells. Nature 414, 105–111 (2001).
Passegue, E., Jamieson, C.H., Ailles, L.E. & Weissman, I.L. Normal and leukemic hematopoiesis: are leukemias a stem cell disorder or a reacquisition of stem cell characteristics? Proc. Natl. Acad. Sci. USA 100 (Suppl.) 11842–11849 (2003).
Hope, K.J., Jin, L. & Dick, J.E. Acute myeloid leukemia originates from a hierarchy of leukemic stem cell classes that differ in self-renewal capacity. Nat. Immunol. 5, 738–743 (2004).
Michor, F. et al. Dynamics of chronic myeloid leukaemia. Nature 435, 1267–1270 (2005).
Wang, J.C. & Dick, J.E. Cancer stem cells: lessons from leukemia. Trends Cell Biol. 15, 494–501 (2005).
Akashi, K., Traver, D., Miyamoto, T. & Weissman, I.L. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature 404, 193–197 (2000).
Manz, M.G., Miyamoto, T., Akashi, K. & Weissman, I.L. Prospective isolation of human clonogenic common myeloid progenitors. Proc. Natl. Acad. Sci. USA 99, 11872–11877 (2002).
Jamieson, C.H. et al. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N. Engl. J. Med. 351, 657–667 (2004).
Steidl, U. et al. Primary human CD34+ hematopoietic stem and progenitor cells express functionally active receptors of neuromediators. Blood 104, 81–88 (2004).
Ivanova, N.B. et al. A stem cell molecular signature. Science 298, 601–604 (2002).
Kiel, M.J. et al. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell 121, 1109–1121 (2005).
Li, C. & Wong, W.H. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA 98, 31–36 (2001).
Li, C. & Hung, W.W. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2 RESEARCH0032 (2001).
Ramalho-Santos, M., Yoon, S., Matsuzaki, Y., Mulligan, R.C. & Melton, D.A. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science 298, 597–600 (2002).
Landgrebe, J., Wurst, W. & Welzl, G. Permutation-validated principal components analysis of microarray data. Genome Biol. 3 RESEARCH0019 (2002).
Donninger, H. et al. Whole genome expression profiling of advance stage papillary serous ovarian cancer reveals activated pathways. Oncogene 23, 8065–8077 (2004).
Kobayashi, S. et al. Calpain-mediated X-linked inhibitor of apoptosis degradation in neutrophil apoptosis and its impairment in chronic neutrophilic leukemia. J. Biol. Chem. 277, 33968–33977 (2002).
Kobayashi, S. et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 352, 786–792 (2005).
Pear, W.S. et al. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood 92, 3780–3792 (1998).
DeKoter, R.P., Walsh, J.C. & Singh, H.P.U. 1 regulates both cytokine-dependent proliferation and differentiation of granulocyte/macrophage progenitors. EMBO J. 17, 4456–4468 (1998).
Steidl, U. et al. Gene expression profiling identifies significant differences between the molecular phenotypes of bone marrow-derived and circulating human CD34+ hematopoietic stem cells. Blood 99, 2037–2044 (2002).
Dolbeare, F., Gratzner, H., Pallavicini, M.G. & Gray, J.W. Flow cytometric measurement of total DNA content and incorporated bromodeoxyuridine. Proc. Natl. Acad. Sci. USA 80, 5573–5577 (1983).
Verhaak, R.G. et al. Mutations in nucleophosmin (NPM1) in acute myeloid leukemia (AML): association with other gene abnormalities and previously established gene expression signatures and their favorable prognostic significance. Blood 106, 3747–3754 (2005).
Acknowledgements
We thank K. Martens for excellent assistance with mouse husbandry, M. Joseph for help with array hybridization, T. Dajaram and C. Hetherington for quantitative real-time RT-PCR analysis and J. Tigges and V. Toxavidis for expert assistance with multicolor flow cytometry and high-speed cell sorting. U.S. thanks S. Steidl for invaluable support and advice. U.S. also thanks R. Kronenwett and R. Haas for long-term support and mentorship. This work was supported by US National Institutes of Health grant CA41456 to D.G.T. and by fellowships of the Dr. Mildred Scheel Foundation for Cancer Research to U.S. (D/03/41221) and the Lymphoma Research Foundation to F.R.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Principal component analysis separates PU.1 knockdown from wild-type HSC on the basis of their gene expresson profiles. (PDF 14 kb)
Supplementary Fig. 2
Gene expression data of wild-type and PU.1 knockdown HSC measured by Affymetrix Mouse Genome 430 2.0 arrays. (PDF 20 kb)
Supplementary Fig. 3
Direct interaction pathway analysis. (PDF 187 kb)
Supplementary Fig. 4
Restoration of c-Jun expression rescues colony formation upon GM-CSF stimulation. (PDF 19 kb)
Supplementary Fig. 5
Flow cytometric analysis shows engraftment of GFP lentivirus-transduced PU.1 knockdown cells upon transplantation into NOD-SCID mice. (PDF 25 kb)
Supplementary Fig. 6
JunB and PU.1 expression in individuals with AML. (PDF 122 kb)
Supplementary Table 1
Selection of up- and downregulated genes in PU.1 knockdown HSC. (PDF 74 kb)
Supplementary Table 2
Oligonucleotides used for PCR and EMSA. (PDF 26 kb)
Rights and permissions
About this article
Cite this article
Steidl, U., Rosenbauer, F., Verhaak, R. et al. Essential role of Jun family transcription factors in PU.1 knockdown–induced leukemic stem cells. Nat Genet 38, 1269–1277 (2006). https://doi.org/10.1038/ng1898
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1898