Abstract
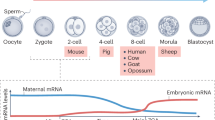
To better understand transcriptional regulation during human oogenesis and preimplantation development, we defined stage-specific transcription, which highlighted the cleavage stage as being highly distinctive. Here, we present multiple lines of evidence that a eutherian-specific multicopy retrogene, DUX4, encodes a transcription factor that activates hundreds of endogenous genes (for example, ZSCAN4, KDM4E and PRAMEF-family genes) and retroviral elements (MERVL/HERVL family) that define the cleavage-specific transcriptional programs in humans and mice. Remarkably, mouse Dux expression is both necessary and sufficient to convert mouse embryonic stem cells (mESCs) into 2-cell-embryo-like ('2C-like') cells, measured here by the reactivation of '2C' genes and repeat elements, the loss of POU5F1 (also known as OCT4) protein and chromocenters, and the conversion of the chromatin landscape (as assessed by transposase-accessible chromatin using sequencing (ATAC–seq)) to a state strongly resembling that of mouse 2C embryos. Thus, we propose mouse DUX and human DUX4 as major drivers of the cleavage or 2C state.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Liu, L. et al. Telomere lengthening early in development. Nat. Cell Biol. 9, 1436–1441 (2007).
Matoba, S. et al. Embryonic development following somatic cell nuclear transfer impeded by persisting histone methylation. Cell 159, 884–895 (2014).
Chung, Y.G. et al. Histone demethylase expression enhances human somatic cell nuclear transfer efficiency and promotes derivation of pluripotent stem cells. Cell Stem Cell 17, 758–766 (2015).
Zalzman, M. et al. Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature 464, 858–863 (2010).
Kalmbach, K., Robinson, L.G. Jr., Wang, F., Liu, L. & Keefe, D. Telomere length reprogramming in embryos and stem cells. BioMed Res. Int. 2014, 925121 (2014).
Macfarlan, T.S. et al. Endogenous retroviruses and neighboring genes are coordinately repressed by LSD1/KDM1A. Genes Dev. 25, 594–607 (2011).
Gifford, W.D., Pfaff, S.L. & Macfarlan, T.S. Transposable elements as genetic regulatory substrates in early development. Trends Cell Biol. 23, 218–226 (2013).
Macfarlan, T.S. et al. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487, 57–63 (2012).
Ishiuchi, T. et al. Early embryonic-like cells are induced by downregulating replication-dependent chromatin assembly. Nat. Struct. Mol. Biol. 22, 662–671 (2015).
Eckersley-Maslin, M.A. et al. MERVL/Zscan4 network activation results in transient genome-wide DNA demethylation of mESCs. Cell Rep. 17, 179–192 (2016).
Choi, Y.J. et al. Deficiency of microRNA miR-34a expands cell fate potential in pluripotent stem cells. Science 355, eaag1927 (2017).
Geng, L.N. et al. DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev. Cell 22, 38–51 (2012).
Young, J.M. et al. DUX4 binding to retroelements creates promoters that are active in FSHD muscle and testis. PLoS Genet. 9, e1003947 (2013).
Gertz, J. et al. Transposase mediated construction of RNA-seq libraries. Genome Res. 22, 134–141 (2012).
Xue, Z. et al. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 500, 593–597 (2013).
Yan, L. et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 20, 1131–1139 (2013).
Leidenroth, A. & Hewitt, J.E. A family history of DUX4: phylogenetic analysis of DUXA, B, C and Duxbl reveals the ancestral DUX gene. BMC Evol. Biol. 10, 364 (2010).
Holland, P.W.H., Booth, H.A.F. & Bruford, E.A. Classification and nomenclature of all human homeobox genes. BMC Biol. 5, 47 (2007).
Bürglin, T.R. & Affolter, M. Homeodomain proteins: an update. Chromosoma 125, 497–521 (2016).
Dunwell, T.L. & Holland, P.W.H. Diversity of human and mouse homeobox gene expression in development and adult tissues. BMC Dev. Biol. 16, 40 (2016).
Madissoon, E. et al. Characterization and target genes of nine human PRD-like homeobox domain genes expressed exclusively in early embryos. Sci. Rep. 6, 28995 (2016).
Töhönen, V. et al. Novel PRD-like homeodomain transcription factors and retrotransposon elements in early human development. Nat. Commun. 6, 8207 (2015).
Jouhilahti, E.-M. et al. The human PRD-like homeobox gene LEUTX has a central role in embryo genome activation. Development 143, 3459–3469 (2016).
Ko, M.S.H. in Mammalian Preimplantation Development 120, 103–124 (Elsevier, 2016).
Göke, J. et al. Dynamic transcription of distinct classes of endogenous retroviral elements marks specific populations of early human embryonic cells. Cell Stem Cell 16, 135–141 (2015).
McLean, C.Y. et al. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 (2010).
Deng, Q., Ramsköld, D., Reinius, B. & Sandberg, R. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science 343, 193–196 (2014).
Leidenroth, A. et al. Evolution of DUX gene macrosatellites in placental mammals. Chromosoma 121, 489–497 (2012).
Clapp, J. et al. Evolutionary conservation of a coding function for D4Z4, the tandem DNA repeat mutated in facioscapulohumeral muscular dystrophy. Am. J. Hum. Genet. 81, 264–279 (2007).
Eidahl, J.O. et al. Mouse Dux is myotoxic and shares partial functional homology with its human paralog DUX4. Hum. Mol. Genet. 25, 4577–4589 (2016).
Schoorlemmer, J., Pérez-Palacios, R., Climent, M., Guallar, D. & Muniesa, P. Regulation of mouse retroelement MuERV-L/MERVL expression by REX1 and epigenetic control of stem cell potency. Front. Oncol. 4, 14 (2014).
Kigami, D., Minami, N., Takayama, H. & Imai, H. MuERV-L is one of the earliest transcribed genes in mouse one-cell embryos. Biol. Reprod. 68, 651–654 (2003).
Ribet, D. et al. Murine endogenous retrovirus MuERV-L is the progenitor of the “orphan” epsilon viruslike particles of the early mouse embryo. J. Virol. 82, 1622–1625 (2008).
Buenrostro, J.D., Giresi, P.G., Zaba, L.C., Chang, H.Y. & Greenleaf, W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Wu, J. et al. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature 534, 652–657 (2016).
Zhou, L.-Q. & Dean, J. Reprogramming the genome to totipotency in mouse embryos. Trends Cell Biol. 25, 82–91 (2015).
Ishiuchi, T. & Torres-Padilla, M.-E. Towards an understanding of the regulatory mechanisms of totipotency. Curr. Opin. Genet. Dev. 23, 512–518 (2013).
Harrison, M.M., Li, X.-Y., Kaplan, T., Botchan, M.R. & Eisen, M.B. Zelda binding in the early Drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition. PLoS Genet. 7, e1002266 (2011).
Sun, Y. et al. Zelda overcomes the high intrinsic nucleosome barrier at enhancers during Drosophila zygotic genome activation. Genome Res. 25, 1703–1714 (2015).
Iwafuchi-Doi, M. & Zaret, K.S. Pioneer transcription factors in cell reprogramming. Genes Dev. 28, 2679–2692 (2014).
Morgani, S.M. & Brickman, J.M. The molecular underpinnings of totipotency. Phil. Trans. R. Soc. Lond. B 369, 20130549 (2014).
De Paepe, C., Krivega, M., Cauffman, G., Geens, M. & Van de Velde, H. Totipotency and lineage segregation in the human embryo. Mol. Hum. Reprod. 20, 599–618 (2014).
Borsos, M. & Torres-Padilla, M.-E. Building up the nucleus: nuclear organization in the establishment of totipotency and pluripotency during mammalian development. Genes Dev. 30, 611–621 (2016).
Yasuda, T. et al. Recurrent DUX4 fusions in B cell acute lymphoblastic leukemia of adolescents and young adults. Nat. Genet. 48, 569–574 (2016).
Zhang, J. et al. Deregulation of DUX4 and ERG in acute lymphoblastic leukemia. Nat. Genet. 48, 1481–1489 (2016).
Patro, R., Mount, S.M. & Kingsford, C. Sailfish enables alignment-free isoform quantification from RNA-seq reads using lightweight algorithms. Nat. Biotechnol. 32, 462–464 (2014).
Niakan, K.K. & Eggan, K. Analysis of human embryos from zygote to blastocyst reveals distinct gene expression patterns relative to the mouse. Dev. Biol. 375, 454–464 (2012).
Bailey, T.L. et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37 (Suppl. 2), W202–W208 (2009).
Buenrostro, J.D., Giresi, P.G., Zaba, L.C., Chang, H.Y. & Greenleaf, W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Ramírez, F., Dündar, F., Diehl, S., Grüning, B.A. & Manke, T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–W191 (2014).
Yu, G., Wang, L.G. & He, Q.Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383 (2015).
Acknowledgements
We thank S. Kuerten (NuGen) for assistance with preparing the RNA-seq libraries, B. Dalley for sequencing services, and T. Parnell for bioinformatic assistance. We give special thanks to M.-E. Torres-Padilla (IGBMC) for generously providing the MERVL::GFP reporter mESC line, and we thank D. Root (Broad Institute) for providing materials. Functional genomics work was supported by HHMI. J.A.D. was further supported by Eunice Kennedy Shriver NIH NICHD K12HD000849. S.J.T. and J.-W.L. were supported by NIH NIAMS R01AR045203, NIH NINDS P01NS069539, and the Friends of FSH Research. J.L.W. was supported by the National Science Foundation Graduate Research Fellowship Program DGE-1256082 and the University of Washington Interdisciplinary Training in Genome Sciences grant T32 HG00035 from NHGRI. Finally, we acknowledge CA042014 for support of the University of Utah core facilities.
Author information
Authors and Affiliations
Contributions
IRB processing, patient consent, patient management, and sample selection/processing were overseen by J.A.D., D.T.C., and C.M.P., with processing by clinical staff (B.R.E. and A.L.W.) in clinical (non-federally funded) facilities. After cDNA and library preparation, subsequent sequencing and transcriptome analyses, along with all molecular and functional approaches were overseen by B.R.C., with contributions from S.J.T. Experiments were performed, analyzed, and statistically evaluated by P.G.H., with contributions from E.J.G., J.L.W., J.-W.L., C.L.W., B.D.W., C.P., B.R.E., and D.A.N. The manuscript was written by P.G.H. and B.R.C.
Corresponding authors
Ethics declarations
Competing interests
B.R.C., E.J.G., P.G.H., J.L.W., and S.J.T. have filed a provisional patent application, ‘Compositions and methods for reprogramming cells and for somatic cell nuclear transfer using DUXC expression’ (US provisional application no. 62/410,078, US Patent and Trademark Office), which is based in part on this work.
Integrated supplementary information
Supplementary Figure 1 Improved RNA-sequencing methods reveal novel transcription, dynamic splice-isoform expression, and stage-specific gene expression in human oocytes and preimplantation development.
(a) Screenshot of the TET3 gene, as an example of a genomic locus displaying read coverage bias in previous single cell datasets (Yan et al., 2013 in green; Xue et al., 2013 in orange). (b) Gene expression correlations using per-stage average FPKM data; r-values were calculated using a spearman rank statistic. S: single cell; P: pooled cells. (c) Bar graphs comparing total exonic transcription (left panel) and novel transcription (right panel) measured in base pairs; employing thresholds of >1, >3 or >5 reads per region. Exonic transcription encompasses all base pairs annotated by Ensemble, UCSC, and NONCODE. (d) Bar chart depicting the number of transcript isoforms expressed by developmental stage. (e) A non-canonical NANOG isoform is expressed specifically in the cleavage stage. (f) A non-canonical TET2 isoform is maternally loaded; producing a severely truncated protein product that excludes both known functional domains [CD-Cys-rich domain; DSBH-Double-stranded β-helix dioxygenase domain]. (g) The top five de novo motifs enriched in cluster 1 (left) and cluster 7 (right) gene promoters after filtering for match score (>0.70). *Note- an OCT/POU-like motif was highly enriched in cluster 7; however, it fell below the score cutoff (0.61).
Supplementary Figure 2 DUX4 directly activates the genes and repeat elements that are transiently expressed during the human cleavage stage.
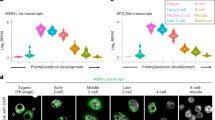
(a) Single cell expression data (RPKM) for DUX4 (RNA-seq data from ref. 16). (b) An arbitrarily rooted phylogenetic tree of human paired (PRD) homeodomains; both homeodomains for the ‘double homeobox’ (DUX) factors are included separately and can be distinguished by the number following the ‘HD’ designation. Orange font indicates genes enriched in the cleavage embryo. Green font is used to delineate mouse DUX homeodomains; the functional ortholog of human DUX4. (c) Single cell expression data (RPKM) for notable double homeobox and ‘PRD-like’ genes (RNA-seq data from ref. 16). (d) The overlap of differentially expressed genes in human iPSCs expressing DUX4 (vs. luciferase) for 14 or 24hrs. (e) Box plot displaying the embryonic expression of the 150 common genes that are upregulated following DUX4 overexpression (for 14hr or 24hrs) in iPSCs (f) MA-plot showing repeat element (by subfamily) activation in human iPSCs 24hrs post DUX4 overexpression (vs luciferase control). (g) The embryonic expression of satellite repeats- HSATII and ACRO1. (h) The overlap of DUX4 ChIP-seq peaks in iPSCs (red) with DUX4 ChIP-seq peaks in myoblasts (MB) from Geng et al., 2012 (light blue). [Overlap statistic calculated by hypergeometric test]. (i) Genome snapshots of cleavage-specific genes directly bound and activated by DUX4 in human iPSCs. (j) The number of repeat element instances uniquely bound by DUX4 for select activated (MLT2A1, MLT2A2, HSATII) and unaffected (LTR7, L1) subfamilies. [Enrichment statistic determined empirically; error bars, s.d.].
Supplementary Figure 3 Mouse Dux, a functional ortholog of DUX4, activates a 2C transcriptional program and converts mESCs to a 2C-like state.
(a) DUX4 and DUX amino acid sequence alignment. Highlighted in blue, green, and yellow are the two DUX4 homeodomains (HD) and the transactivation domain (TAD), respectively. (b) RT-qPCR data for select ‘2C’ genes activated following Dux expression in mouse C2C12 cells [three replicates per condition. Error bars, s.d.]. (c) Results of a live imaging experiment showing the relative gain of GFPpos cells (normalized by total cell surface area) as a function of time post dox-induction. (d) Schematic of the RNA-seq experiments conducted on Dux-expressing mESCs. (e) Overlap of differentially expressed genes (DEGs) from unsorted and sorted populations of Dux-expressing mESCs [Overlap statistic calculated by hypergeometric test]. (f) The normalized average expression of codon altered Dux transgene in our RNA-seq datasets from unsorted and sorted populations (left panel), relative to the normalized expression of endogenous Dux in spontaneously converting 2C-like cells (right panel) (RNA-seq data from ref. 9). (g) MA-plot showing the activation of repetitive elements (by subfamily) in both unsorted and sorted RNA-seq experiments. Notably, Dux expression robustly induces the expression of MERVL elements and pericentromeric major satellite repeats (GSAT). (h) Flow results demonstrating, in an independent HA-tagged clone, the ability of Dux expression to efficiently induce reactivation of the MERVL reporter in mESCs [three biological replicates per condition; error bars, s.d.]. (i) The expression of HA and loss of chromocenters is evaluated by immunofluorescence confirming entry into a 2C-like state. Scale bar, 10um.
Supplementary Figure 4 Dux is necessary for spontaneous and CAF-1-mediated conversion of mESCs to a 2C-like state.
(a) A diagram of the Chromatin Assemble Factor (CAF-1) complex. The arrow points to the complex subunit (p150 encoded by the Chaf1a gene) targeted with siRNAs in our experiments. (b) Dot plot depicting the correlation of gene expression changes in the Dux-induced 2C-like cells, and those induced by Chaf1a knockdown (RNA-seq data from ref. 9). (c) Effects of Dux knockdown alone (left panel) and Chaf1a knockdown alone (right panel) on conversion of mESCs to a 2C-like state [three biological replicates per condition. Statistics determined using a two-tailed unpaired t-test, error bars, s.d.]. (d) The normalized average expression of Chaf1a and Dux in negative control (NC) and knockdown mESCs determined by RNA-seq [Error bars, s.d.]. (e) Bar chart showing the fraction of genes upregulated (FC>2, FDR<0.01) in Chaf1a depleted mESCs that are not affected in mESCs depleted for both Chaf1a and Dux. (note: one gene that was upregulated in Chaf1a depleted mESCs became downregulated in mESCs depleted for both Chaf1a and Dux). (f) The normalized average expression of MERVL-int and GSAT repeats in control and knockdown mESCs determined by RNA-seq [Error bars, s.d.]. (g) Screenshots showing the expression of notable genes following knockdown of Chaf1a alone and in combination with knockdown of Dux. (h) Boxplot showing the embryonic expression of the genes upregulated in both Chaf1a-depleted as well as Chaf1a- and Dux-depleted mESCs (termed ‘Dux-independent’) and the genes upregulated only in Chaf1a-depleted cells (termed ‘Dux-dependent’). Center line, median; box limits, upper and lower quartiles; whiskers, 1.5× interquartile range. (RNA-seq data from ref. 27) (i) Summary figure depicting the proposed relationship between CAF-1 and DUX with respect to mESC entry into a 2C-like state.
Supplementary Figure 5 Dux-induced 2C-like cells acquire an open chromatin landscape resembling that of an early 2-cell-stage embryo.
(a) Heatmap depicting the Pearson correlation of genome-wide ATAC-seq coverage profiles in Dux-induced mESCs and early embryonic developmental stages (Embryo ATAC-seq data from ref. 35). (b) Pie charts depicting the distribution of ATAC-seq gained, lost and common peaks (called after filtering alignment files for unique reads only) at basic genomic features. Inset pie charts indicate the percentage of unique peaks which overlap with MERVL elements (MT2_Mm and MERVL-int). [Enrichment statistic determined empirically]. (c) Boxplot shows the median log2 expression fold change (FC) of the genes neighboring regions of ATAC-seq gained, lost and common signal.
Supplementary Figure 6 DUX binds directly to 2C gene promoters and retrotransposons.
(a) Heatmap depicting gene clusters exhibiting stage-specific expression in the early mouse embryo (left panel). Overlap of DUX-ChIP occupied genes with each ‘stage-specific’ gene cluster (right panel) [overlap statistics determined by hypergeometric test]. (b) The number of repeat element instances uniquely bound by DUX for select affected (MT2_Mm, ORR1A3-int) and unaffected (L1, IAPEZ-int) subfamilies [enrichment statistics determined empirically; error bars, s.d.]. (c) The percentage of unique ATAC gained, lost, and common regions bound by DUX. (d) A binding motif for DUX predicted by MEME-ChIP based on the top 10,000 peak summits (left panel). This motif differs from that for DUX4, and only shows enrichment in mouse-specific regions of interest (right panel).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 (PDF 1706 kb)
Supplementary Table 1
Egg/Embryo RNA-Seq quality control - Read depth (sheet 1) and quality control metrics (sheet 2) of human oocyte and embryo RNA sequencing. (XLSX 54 kb)
Supplementary Table 2
Egg/Embryo RNA-seq gene and repeat expression- Full expression analysis of all genes (sheet1) and repetitive elements (sheet 2) in the human oocytes and embryos. (XLSX 12882 kb)
Supplementary Table 3
Egg/Embryo heatmap- A list of all 9,734 ensembl genes comprising the heatmap in Fig.1 with cluster and FPKM information. (XLSX 1744 kb)
Supplementary Table 4
Egg/Embryo novel transcription- Full list of all novel transcription fragments (transfrags) and their differential expression. (XLSX 20735 kb)
Supplementary Table 5
Egg/Embryo isoform expression- Expression data (TPM) for all ensembl gene isoforms in human oocytes/embryos estimated by Sailfish (Patro et al.,2014). (XLSX 19464 kb)
Supplementary Table 6
RNA-seq in iPSCs - Full expression analysis of all genes (sheet1) and repetitive elements (sheet 2) in human induced pluripotent stem cells (iPSCs) following hDUX4 or luciferase expression. (XLSX 8683 kb)
Supplementary Table 7
hDUX4 ChIP-seq in iPSCs- All hDUX4 ChIP-seq peaks in iPSCs (qval<10-20) called by MACS2 (over hDUX4 ChIP control) for replicates 1 (sheet 1) and 2 (sheet 2). (XLSX 10165 kb)
Supplementary Table 8
RNA-seq in non-clonal mESCs- Full expression analysis of all genes (sheet1) and repetitive elements (sheet 2) in non-clonal mESCs post transient mDux expression. (XLSX 3637 kb)
Supplementary Table 9
RNA-seq in clonal mESCs- Full expression analysis of all genes (sheet1) and repetitive elements (sheet 2) in a clonal, unsorted population of mESCs plus/minus 24hrs of dox-inducible mDux expression. (XLSX 3865 kb)
Supplementary Table 10
RNA-seq in sorted '2C-like' cells- Full expression analysis of all genes (sheet1) and repetitive elements (sheet 2) in a clonal, sorted (GFPpos and GFPneg) population of mESCs after 24hrs of doxinducible mDux expression. (XLSX 3968 kb)
Supplementary Table 11
RNA-seq in siRNA-treated mESCs- Full expression analysis of all genes (sheet1) and repetitive elements (sheet 2) in mESCs treated with siRNAs against Chaf1a alone or in combination with siRNAs against mDux (si308 and si309). (XLSX 7564 kb)
Supplementary Table 12
ATAC-seq in sorted '2C-like' cells- a list of regions pertaining to the MACS2 ATAC-seq peaks gained in GFPpos mESCs (sheet 1), lost in GFPpos mESCs (sheet 2), or found in common between GFPpos and GFPneg mESCs (sheet3) induced with 24hrs of mDux expression. (XLSX 934 kb)
Supplementary Table 13
mDUX ChIP-seq in mESCs- All mDUX ChIP-seq peaks in mESCs (qval<0.05]) called by MACS2 (over input) for replicates 1 (sheet 1) and 2 (sheet 2). (XLSX 3867 kb)
Supplementary Table 14
Primer sequences- A list of all primers used for RT-qPCR experiments (sheet 1) and generation of siRNA pools (sheet 2). (XLSX 9 kb)
Rights and permissions
About this article
Cite this article
Hendrickson, P., Doráis, J., Grow, E. et al. Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat Genet 49, 925–934 (2017). https://doi.org/10.1038/ng.3844
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3844