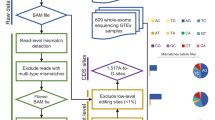
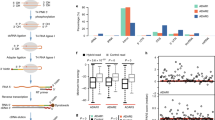
Abstract
Adenosine-to-inosine (A-to-I) RNA editing is a post-transcriptional processing event involved in diversifying the transcriptome responsible for various biological processes. Although bioinformatic approaches have predicted a number of A-to-I editing sites in cDNAs, the human transcriptome is thought to still harbor large numbers of as-yet-unknown editing sites. Exploring new editing sites requires a biochemical method to accurately identify inosines on RNA strands. We here describe a chemical method to identify inosines, called inosine chemical erasing (ICE), that is based on cyanoethylation combined with reverse transcription. We carried out a large-scale verification of the ICE method focusing on 642 regions in human cDNA and identified 5,072 editing sites, including 4,395 new sites. Functional study revealed that A-to-I intronic editing in the SARS gene mediated by ADAR1 is involved in preventing aberrant exonization of Alu sequence into mature mRNA.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Bass, B.L. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 71, 817–846 (2002).
Higuchi, M. et al. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 406, 78–81 (2000).
Wang, Q., Khillan, J., Gadue, P. & Nishikura, K. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science 290, 1765–1768 (2000).
Wang, Q. et al. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J. Biol. Chem. 279, 4952–4961 (2004).
Jepson, J.E. & Reenan, R.A. RNA editing in regulating gene expression in the brain. Biochim. Biophys. Acta 1779, 459–470 (2008).
Maas, S., Kawahara, Y., Tamburro, K.M. & Nishikura, K. A-to-I RNA editing and human disease. RNA Biol. 3, 1–9 (2006).
Maas, S., Patt, S., Schrey, M. & Rich, A. Underediting of glutamate receptor GluR-B mRNA in malignant gliomas. Proc. Natl. Acad. Sci. USA 98, 14687–14692 (2001).
Kawahara, Y. et al. Glutamate receptors: RNA editing and death of motor neurons. Nature 427, 801 (2004).
Levanon, E.Y. et al. Evolutionarily conserved human targets of adenosine to inosine RNA editing. Nucleic Acids Res. 33, 1162–1168 (2005).
Levanon, E.Y. et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 22, 1001–1005 (2004).
Kim, D.D. et al. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 14, 1719–1725 (2004).
Athanasiadis, A., Rich, A. & Maas, S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2, e391 (2004).
Levanon, K., Eisenberg, E., Rechavi, G. & Levanon, E.Y. Letter from the editor: Adenosine-to-inosine RNA editing in Alu repeats in the human genome. EMBO Rep. 6, 831–835 (2005).
Hoopengardner, B., Bhalla, T., Staber, C. & Reenan, R. Nervous system targets of RNA editing identified by comparative genomics. Science 301, 832–836 (2003).
Burns, C.M. et al. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature 387, 303–308 (1997).
Higuchi, M. et al. RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency. Cell 75, 1361–1370 (1993).
Rueter, S.M., Dawson, T.R. & Emeson, R.B. Regulation of alternative splicing by RNA editing. Nature 399, 75–80 (1999).
Chen, L.L., DeCerbo, J.N. & Carmichael, G.G. Alu element-mediated gene silencing. EMBO J. 27, 1694–1705 (2008).
Agranat, L., Raitskin, O., Sperling, J. & Sperling, R. The editing enzyme ADAR1 and the mRNA surveillance protein hUpf1 interact in the cell nucleus. Proc. Natl. Acad. Sci. USA 105, 5028–5033 (2008).
Bass, B.L. How does RNA editing affect dsRNA-mediated gene silencing? Cold Spring Harb. Symp. Quant. Biol. 71, 285–292 (2006).
Osenberg, S., Dominissini, D., Rechavi, G. & Eisenberg, E. Widespread cleavage of A-to-I hyperediting substrates. RNA 15, 1632–1639 (2009).
Hundley, H.A., Krauchuk, A.A. & Bass, B.L. C. elegans and H. sapiens mRNAs with edited 3′ UTRs are present on polysomes. RNA 14, 2050–2060 (2008).
Borchert, G.M. et al. Adenosine deamination in human transcripts generates novel microRNA binding sites. Hum. Mol. Genet. 18, 4801–4807 (2009).
Nishikura, K. Editor meets silencer: crosstalk between RNA editing and RNA interference. Nat. Rev. Mol. Cell Biol. 7, 919–931 (2006).
Kawahara, Y. et al. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science 315, 1137–1140 (2007).
Paz, N. et al. Altered adenosine-to-inosine RNA editing in human cancer. Genome Res. 17, 1586–1595 (2007).
Nishimoto, Y. et al. Determination of editors at the novel A-to-I editing positions. Neurosci. Res. 61, 201–206 (2008).
Yoshida, M. & Ukita, T. Modification of nucleosides and nucleotides. VII. Selective cyanoethylation of inosine and pseudouridine in yeast transfer ribonucleic acid. Biochim. Biophys. Acta 157, 455–465 (1968).
Mengel-Jørgensen, J. & Kirpekar, F. Detection of pseudouridine and other modifications in tRNA by cyanoethylation and MALDI mass spectrometry. Nucleic Acids Res. 30, e135 (2002).
Chambers, R.W. The chemistry of pseudouridine. IV. cyanoethylation. Biochemistry 4, 219–226 (1965).
Chambers, R.W., Kurkov, V. & Shapiro, R. The chemistry of pseudouridine. synthesis of pseudouridine-5′-diphosphate. Biochemistry 2, 1192–1203 (1963).
Yoshida, M. & Ukita, T. Modification of nucleosides and nucleotides. 8. The reaction rates of pseudouridine residues with acrylonitrile and its relation to the secondary structure of transfer ribonucleic acid. Biochim. Biophys. Acta 157, 466–475 (1968).
Seeburg, P.H., Higuchi, M. & Sprengel, R. RNA editing of brain glutamate receptor channels: mechanism and physiology. Brain Res. Brain Res. Rev. 26, 217–229 (1998).
Sherry, S.T. et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29, 308–311 (2001).
Kawahara, Y. et al. Frequency and fate of microRNA editing in human brain. Nucleic Acids Res. 36, 5270–5280 (2008).
Frommer, M. et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. USA 89, 1827–1831 (1992).
Schaefer, M., Pollex, T., Hanna, K. & Lyko, F. RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res. 37, e12 (2009).
Hartner, J.C. et al. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J. Biol. Chem. 279, 4894–4902 (2004).
Hartner, J.C., Walkley, C.R., Lu, J. & Orkin, S.H. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat. Immunol. 10, 109–115 (2009).
George, C.X., Wagner, M.V. & Samuel, C.E. Expression of interferon-inducible RNA adenosine deaminase ADAR1 during pathogen infection and mouse embryo development involves tissue-selective promoter utilization and alternative splicing. J. Biol. Chem. 280, 15020–15028 (2005).
Patterson, J.B. & Samuel, C.E. Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: evidence for two forms of the deaminase. Mol. Cell. Biol. 15, 5376–5388 (1995).
Ohlson, J., Enstero, M., Sjoberg, B.M. & Ohman, M. A method to find tissue-specific novel sites of selective adenosine deamination. Nucleic Acids Res. 33, e167 (2005).
Morse, D.P. & Bass, B.L. Detection of inosine in messenger RNA by inosine-specific cleavage. Biochemistry 36, 8429–8434 (1997).
Sakurai, M., Ohtsuki, T., Suzuki, T. & Watanabe, K. Unusual usage of wobble modifications in mitochondrial tRNAs of the nematode Ascaris suum. FEBS Lett. 579, 2767–2772 (2005).
Suzuki, T., Suzuki, T., Wada, T., Saigo, K. & Watanabe, K. Taurine as a constituent of mitochondrial tRNAs: new insights into the functions of taurine and human mitochondrial diseases. EMBO J. 21, 6581–6589 (2002).
Sprinzl, M., Horn, C., Brown, M., Ioudovitch, A. & Steinberg, S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 26, 148–153 (1998).
Crooks, G.E., Hon, G., Chandonia, J.M. & Brenner, S.E. WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 (2004).
Acknowledgements
We are grateful to the members of the Suzuki laboratory, especially to S. Okada and to H. Terajima for his experimental assistance and fruitful discussions on this study. Special thanks are due to Maze, Inc. for their support of computational work. This work was supported by Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture of Japan and by a grant from the New Energy and Industrial Technology Development Organization (to T. S.).
Author information
Authors and Affiliations
Contributions
M.S. developed and optimized the ICE method. T.Y. performed functional analyses of the identified sites. H.K. performed a genome-wide identification of the editing sites. H.U. carried out all computational work. T.S. designed and supervised all the work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Methods, Supplementary Figures 1–12 and Supplementary Tables 1 & 2 (PDF 4147 kb)
Supplementary Tables
Supplementary Tables 3–6 (XLS 1317 kb)
Rights and permissions
About this article
Cite this article
Sakurai, M., Yano, T., Kawabata, H. et al. Inosine cyanoethylation identifies A-to-I RNA editing sites in the human transcriptome. Nat Chem Biol 6, 733–740 (2010). https://doi.org/10.1038/nchembio.434
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.434