Abstract
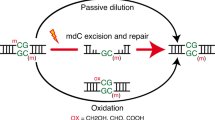
5-Hydroxymethylcytosine (hmC) is an oxidation product of 5-methylcytosine which is present in the deoxyribonucleic acid (DNA) of most mammalian cells. Reduction of hmC levels in DNA is a hallmark of cancers. Elucidating the dynamics of this oxidation reaction and the lifetime of hmC in DNA is fundamental to understanding hmC function. Using stable isotope labelling of cytosine derivatives in the DNA of mammalian cells and ultrasensitive tandem liquid–chromatography mass spectrometry, we show that the majority of hmC is a stable modification, as opposed to a transient intermediate. In contrast with DNA methylation, which occurs immediately during replication, hmC forms slowly during the first 30 hours following DNA synthesis. Isotopic labelling of DNA in mouse tissues confirmed the stability of hmC in vivo and demonstrated a relationship between global levels of hmC and cell proliferation. These insights have important implications for understanding the states of chemically modified DNA bases in health and disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Klose, R. J. & Bird, A. P. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 31, 89–97 (2006).
Kriaucionis, S. & Heintz, N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324, 929–930 (2009).
Tahiliani, M. et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 (2009).
Globisch, D. et al. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS ONE 5, e15367 (2010).
Haffner, M. C. et al. Global 5-hydroxymethylcytosine content is significantly reduced in tissue stem/progenitor cell compartments and in human cancers. Oncotarget 2, 627–637 (2011).
Jin, S. G. et al. 5-Hydroxymethylcytosine is strongly depleted in human cancers but its levels do not correlate with IDH1 mutations. Cancer Res. 71, 7360–7365 (2011).
Lian, C. G. et al. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell 150, 1135–1146 (2012).
Kraus, T. F. et al. Low values of 5-hydroxymethylcytosine (5hmC), the “sixth base,” are associated with anaplasia in human brain tumors. Int. J. Cancer 131, 1577–1590 (2012).
Ito, S. et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466, 1129–1133 (2010).
He, Y. F. et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307 (2011).
Ito, S. et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303 (2011).
Song, C. X. & He, C. Potential functional roles of DNA demethylation intermediates. Trends Biochem. Sci. 38, 480–484 (2013).
Mellen, M., Ayata, P., Dewell, S., Kriaucionis, S. & Heintz, N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell 151, 1417–1430 (2012).
Iurlaro, M. et al. A screen for hydroxymethylcytosine and formylcytosine binding proteins suggests functions in transcription and chromatin regulation. Genome Biol. 14, R119 (2013).
Spruijt, C. G. et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell 152, 1146–1159 (2013).
Yu, M. et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 149, 1368–1380 (2012).
Williams, K. et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 473, 343–348 (2011).
Wu, H. et al. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 25, 679–684 (2011).
Xu, Y. et al. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol. Cell 42, 451–464 (2011).
Shen, L. & Zhang, Y. 5-Hydroxymethylcytosine: generation, fate, and genomic distribution. Curr. Opin. Cell Biol. 25, 289–296 (2013).
Merrill, G. F. Cell synchronization. Methods Cell Biol. 57, 229–249 (1998).
Kappler, J. W. The kinetics of DNA methylation in cultures of a mouse adrenal cell line. J. Cell Physiol. 75, 21–31 (1970).
Woodcock, D. M. et al. Delayed DNA methylation is an integral feature of DNA replication in mammalian cells. Exp. Cell Res. 166, 103–112 (1986).
Otani, J. et al. Cell cycle-dependent turnover of 5-hydroxymethyl cytosine in mouse embryonic stem cells. PLoS ONE 8, e82961 (2013).
Hansen, R. S. et al. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc. Natl Acad. Sci. USA 107, 139–44 (2010).
Vassilev, L. & Russev, G. Purification of nascent DNA chains by immunoprecipitation with anti-BrdU antibodies. Nucleic Acids Res. 16, 10397 (1988).
Guo, J. U., Su, Y., Zhong, C., Ming, G. L. & Song, H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell 145, 423–434 (2011).
Zhang, R. R. et al. Tet1 regulates adult hippocampal neurogenesis and cognition. Cell Stem Cell 13, 237–245 (2013).
Dawlaty, M. M. et al. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell 9, 166–175 (2011).
Inoue, A. & Zhang, Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science 334, 194 (2011).
Iqbal, K., Jin, S. G., Pfeifer, G. P. & Szabo, P. E. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc. Natl Acad. Sci. USA 108, 3642–3647 (2011).
Salvaing, J. et al. 5-Methylcytosine and 5-hydroxymethylcytosine spatiotemporal profiles in the mouse zygote. PLoS ONE 7, e38156 (2012).
Kohli, R. M. & Zhang, Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature 502, 472–479 (2013).
Wang, L. et al. Programming and inheritance of parental DNA methylomes in mammals. Cell 157, 979–991 (2014).
Minor, E. A., Court, B. L., Young, J. I. & Wang, G. Ascorbate induces ten–eleven translocation (Tet) methylcytosine dioxygenase-mediated generation of 5-hydroxymethylcytosine. J. Biol. Chem. 288, 13669–13674 (2013).
Blaschke, K. et al. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature 500, 222–226 (2013).
Huh, Y. H., Cohen, J. & Sherley, J. L. Higher 5-hydroxymethylcytosine identifies immortal DNA strand chromosomes in asymmetrically self-renewing distributed stem cells. Proc. Natl Acad. Sci. USA 110, 16862–1687 (2013).
Booth, M. J. et al. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science 336, 934–937 (2012).
Ying, Q. L. et al. The ground state of embryonic stem cell self-renewal. Nature 453, 519–523 (2008).
Acknowledgements
We acknowledge the CRUK CI Flow Cytometry and Histopathology/In Situ Hybridization core facilities for their contributions, D. Oxley, C. d'Santos and D. Michelle-Smith for their support with mass spectrometry, X. Zou for his help with mES cells and D. Tannahill for a critical reading of the manuscript. This work was funded by CRUK (all authors) and the Wellcome Trust Senior Investigator Award (S.B.).
Author information
Authors and Affiliations
Contributions
M.B., S.U-L., A.M. and S.B. conceived and designed the experiments; M.B. and S.U-L. performed the experiments; X.Y., M.W. and M.B. developed the mass spectrometry methods; M.B. and S.U-L. analysed the data with the help of X.Y. and M.W.; M.B., S.U-L. and S.B. co-wrote the manuscript with contributions from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 7279 kb)
Rights and permissions
About this article
Cite this article
Bachman, M., Uribe-Lewis, S., Yang, X. et al. 5-Hydroxymethylcytosine is a predominantly stable DNA modification. Nature Chem 6, 1049–1055 (2014). https://doi.org/10.1038/nchem.2064
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchem.2064
This article is cited by
-
Modeling methyl-sensitive transcription factor motifs with an expanded epigenetic alphabet
Genome Biology (2024)
-
Obesity and dyslipidemia are associated with partially reversible modifications to DNA hydroxymethylation of apoptosis- and senescence-related genes in swine adipose-derived mesenchymal stem/stromal cells
Stem Cell Research & Therapy (2023)
-
Genome-wide hydroxymethylation profiles in liver of female Nile tilapia with distinct growth performance
Scientific Data (2023)
-
Impact of antigen retrieval protocols on the immunohistochemical detection of epigenetic DNA modifications
Histochemistry and Cell Biology (2023)
-
Developmental and Injury-induced Changes in DNA Methylation in Regenerative versus Non-regenerative Regions of the Vertebrate Central Nervous System
BMC Genomics (2022)