Abstract
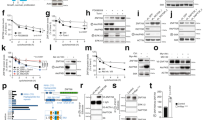
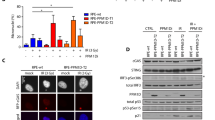
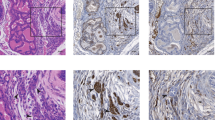
Primary fibroblasts respond to activated H-RASV12 by undergoing premature arrest, which resembles replicative senescence1. This irreversible 'fail-safe mechanism' requires p19ARF, p53 and the Retinoblastoma (Rb) family: upon their disruption, RASV12-expressing cells fail to undergo senescence and continue to proliferate1,2,3,4,5,6,7. Similarly, co-expression of oncogenes such as c-MYC or E1A rescues RASV12-induced senescence. To identify novel genes that allow escape from RASV12-induced senescence, we designed an unbiased, retroviral complementary DNA library screen. We report on the identification of DRIL1, the human orthologue of the mouse Bright and Drosophila dead ringer transcriptional regulators. DRIL1 renders primary murine fibroblasts unresponsive to RASV12-induced anti-proliferative signalling by p19ARF/p53/p21CIP1, as well as by p16INK4a. In this way, DRIL1 not only rescues RASV12-induced senescence but also causes these fibroblasts to become highly oncogenic. Furthermore, DRIL1 immortalizes mouse fibroblasts, in the presence of high levels of p16INK4a. Immortalization by DRIL1, whose product binds the pRB-controlled transcription factor E2F1 (ref. 8), is correlated with induction of E2F1 activity. Correspondingly, DRIL1 induces the E2F1 target Cyclin E1, overexpression of which is sufficient to trigger escape from senescence. Thus, DRIL1 disrupts cellular protection against RASV12-induced proliferation downstream of the p19ARF/p53 pathway.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Serrano, M., Lin, A. W., McCurrach, M. E., Beach, D. & Lowe, S. W. Cell 88, 593–602 (1997).
Kamijo, T. et al. Cell 91, 649–659 (1997).
Palmero, I., Pantoja, C. & Serrano, M. Nature 395, 125–126 (1998).
Sherr, C. J. Genes Dev. 12, 2984–2991 (1998).
Tanaka, N. et al. Cell 77, 829–839 (1994).
Sage, J. et al. Genes Dev. 14, 3037–3050 (2000).
Peeper, D. S., Dannenberg, J. H., Douma, S., te Riele, H. & Bernards, R. Nature Cell Biol. 3, 198–203 (2001).
Suzuki, M. et al. Oncogene 17, 853–865 (1998).
Lee, G. H., Ogawa, K. & Drinkwater, N. R. Am. J. Pathol. 147, 1811–1822 (1995).
Kortschak, R. D. et al. Genomics 51, 288–292 (1998).
Herrscher, R. F. et al. Genes Dev. 9, 3067–3082 (1995).
Gregory, S. L., Kortschak, R. D., Kalionis, B. & Saint, R. Mol. Cell. Biol. 16, 792–799 (1996).
Kortschak, R. D., Tucker, P. W. & Saint, R. Trends Biochem. Sci. 25, 294–299 (2000).
Jacobs, J. J. et al. Nature Genet. 26, 291–299 (2000).
Zindy, F. et al. Genes Dev. 12, 2424–2433 (1998).
Pantoja, C. & Serrano, M. Oncogene 18, 4974–4982 (1999).
Randle, D. H., Zindy, F., Sherr, C. J. & Roussel, M. F. Proc. Natl Acad. Sci. USA 98, 9654–9659 (2001).
Lundberg, A. S. & Weinberg, R. A. Mol. Cell. Biol. 18, 753–761 (1998).
Haas, K. et al. Oncogene 15, 2615–2623 (1997).
Karsunky, H. et al. Oncogene 18, 7816–7824 (1999).
Keyomarsi, K. & Herliczek, T. W. Prog. Cell Cycle Res. 3, 171–191 (1997).
Dannenberg, J. H., van Rossum, A., Schuijff, L. & te Riele, H. Genes Dev. 14, 3051–3064 (2000).
Ohtani, K., DeGregori, J. & Nevins, J. R. Proc. Natl. Acad. Sci. USA 92, 12146–12150 (1995).
Dimri, G. P., Itahana, K., Acosta, M. & Campisi, J. Mol. Cell. Biol. 20, 273–285 (2000).
Johnson, D. G., Cress, W. D., Jakoi, L. & Nevins, J. R. Proc. Natl Acad. Sci. USA 91, 12823–12827 (1994).
Pierce, A. M., Fisher, S. M., Conti, C. J. & Johnson, D. G. Oncogene 16, 1267–1276 (1998).
Lukas, J., Petersen, B. O., Holm, K., Bartek, J. & Helin, K. Mol. Cell. Biol. 16, 1047–1057 (1996).
Alevizopoulos, K., Vlach, J., Hennecke, S. & Amati, B. EMBO J. 16, 5322–5333 (1997).
O'Hagan, R. C. et al. Genes Dev. 14, 2185–2191 (2000).
Groth, A., Weber, J. D., Willumsen, B. M., Sherr, C. J. & Roussel, M. F. J. Biol. Chem. 275, 27473–27480 (2000).
Acknowledgements
We thank C. Sherr for generously providing ARF−/− MEFs, N. Drinkwater for the ts T-antigen plasmid, P. Sicinski for Cyclin D1−/− MEFs, G. Nolan for retroviral packaging cells, T. van Wezel for invaluable help with PCR, J. van der Sman for help with analysis of the screen, H. Starreveld and the animal facility for help with oncogenicity assays, P. Tucker for reagents and for sharing unpublished observations, our colleagues for helpful discussions, and A. Berns, R. Agami, R. Kortlever and B. Rowland for reading the manuscript critically. This work was supported by grants from the Dutch Cancer Society, the Human Frontiers Science Program (HFSP) and the National Institutes of Health (USA). G.D. is the Birnbaum Scholar of the Leukemia and Lymphoma Society of America and a recipient of a Burroughs Wellcome Career Award in the Biomedical Sciences.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Peeper, D., Shvarts, A., Brummelkamp, T. et al. A functional screen identifies hDRIL1 as an oncogene that rescues RAS-induced senescence. Nat Cell Biol 4, 148–153 (2002). https://doi.org/10.1038/ncb742
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb742
This article is cited by
-
RBM4 dictates ESCC cell fate switch from cellular senescence to glutamine-addiction survival through inhibiting LKB1-AMPK-axis
Signal Transduction and Targeted Therapy (2023)
-
ARID3a from the ARID family: structure, role in autoimmune diseases and drug discovery
Acta Pharmacologica Sinica (2023)
-
Hepatic ARID3A facilitates liver cancer malignancy by cooperating with CEP131 to regulate an embryonic stem cell-like gene signature
Cell Death & Disease (2022)
-
Caveolin-1, a master regulator of cellular senescence
Cancer and Metastasis Reviews (2020)
-
Systems-level effects of ectopic galectin-7 reconstitution in cervical cancer and its microenvironment
BMC Cancer (2016)