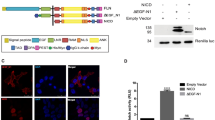
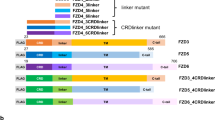
Abstract
Although the Notch and JAK–STAT signalling pathways fulfill overlapping roles in growth and differentiation regulation, no coordination mechanism has been proposed to explain their relationship. Here we show that STAT3 is activated in the presence of active Notch, as well as the Notch effectors Hes1 and Hes5. Hes proteins associate with JAK2 and STAT3, and facilitate complex formation between JAK2 and STAT3, thus promoting STAT3 phosphorylation and activation. Furthermore, suppression of endogenous Hes1 expression reduces growth factor induction of STAT3 phosphorylation. STAT3 seems to be essential for maintenance of radial glial cells and differentiation of astrocytes by Notch in the developing central nervous system. These results suggest that direct protein–protein interactions coordinate cross-talk between the Notch–Hes and JAK–STAT pathways.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Artavanis-Tsakonas, S., Rand, M.D. & Lake, R.J. Notch signaling: cell fate control and signal integration in development. Science 284, 770–776 (1999).
Gaiano, N. & Fishell, G. The role of notch in promoting glial and neural stem cell fates. Annu. Rev. Neurosci. 25, 471–490 (2002).
Weinmaster, G. Notch signal transduction: a real rip and more. Curr. Opin. Genet. Dev. 10, 363–369 (2000).
Levy, D.E. & Darnell, J.E. Jr. Stats: transcriptional control and biological impact. Nature Rev. Mol. Cell. Biol. 3, 651–662 (2002).
Ihle, J.N. The Stat family in cytokine signaling. Curr. Opin. Cell Biol. 13, 211–217 (2001).
Levy, D.E. & Lee, C.K. What does Stat3 do? J. Clin. Invest. 109, 1143–1148 (2002).
Benekli, M., Baer, M.R., Baumann, H. & Wetzler, M. Signal transducer and activator of transcription proteins in leukemias. Blood 101, 2940–2954 (2003).
Radtke, F. & Raj, K. The role of Notch in tumorigenesis: oncogene or tumour suppressor? Nature Rev. Cancer 3, 756–767 (2003).
Chambers, C.B. et al. Spatiotemporal selectivity of response to Notch1 signals in mammalian forebrain precursors. Development 128, 689–702 (2001).
Tanigaki, K. et al. Notch1 and Notch3 instructively restrict bFGF-responsive multipotent neural progenitor cells to an astroglial fate. Neuron 29, 45–55 (2001).
Bonni, A. et al. Regulation of gliogenesis in the central nervous system by the JAK-STAT signaling pathway. Science 278, 477–483 (1997).
Rajan, P. & McKay, R.D. Multiple routes to astrocytic differentiation in the CNS. J. Neurosci. 18, 3620–3629 (1998).
Nakashima, K. et al. Synergistic signaling in fetal brain by STAT3-Smad1 complex bridged by p300. Science 284, 479–482 (1999).
Ge, W. et al. Notch signaling promotes astrogliogenesis via direct CSL-mediated glial gene activation. J. Neurosci. Res. 69, 848–860 (2002).
Yamamoto, N. et al. Role of Deltex-1 as a transcriptional regulator downstream of the Notch receptor. J. Biol. Chem. 276, 45031–45040 (2001).
Hocke, G.M., Barry, D. & Fey, G.H. Synergistic action of interleukin-6 and glucocorticoids is mediated by the interleukin-6 response element of the rat α2 macroglobulin gene. Mol. Cell. Biol. 12, 2282–2294 (1992).
Nakagawa, Y. et al. Roles of cell-autonomous mechanisms for differential expression of region-specific transcription factors in neuroepithelial cells. Development 122, 2449–2464 (1996).
Nakajima, K. et al. A central role for Stat3 in IL-6-induced regulation of growth and differentiation in M1 leukemia cells. EMBO J. 15, 3651–3658 (1996).
Martys-Zage, J.L. et al. Requirement for presenilin 1 in facilitating Jagged 2-mediated endoproteolysis and signaling of notch 1. J. Mol. Neurosci. 15, 189–204 (2000).
Ohtsuka, T. et al. Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J. 18, 2196–2207 (1999).
Kageyama, R. & Nakanishi, S. Helix-loop-helix factors in growth and differentiation of the vertebrate nervous system. Curr. Opin. Genet. Dev. 7, 659–665 (1997).
Satow, T. et al. The basic helix-loop-helix gene hesr2 promotes gliogenesis in mouse retina. J. Neurosci. 21, 1265–1273 (2001).
Hanke, J.H. et al. Discovery of a novel, potent, and Src family-selective tyrosine kinase inhibitor. Study of Lck- and FynT-dependent T cell activation. J. Biol. Chem. 271, 695–701 (1996).
Meydan, N. et al. Inhibition of acute lymphoblastic leukaemia by a Jak-2 inhibitor. Nature 379, 645–648 (1996).
Lobie, P.E. et al. Constitutive nuclear localization of Janus kinases 1 and 2. Endocrinology 137, 4037–4045 (1996).
Ram, P.A. & Waxman, D.J. Interaction of growth hormone-activated STATs with SH2-containing phosphotyrosine phosphatase SHP-1 and nuclear JAK2 tyrosine kinase. J. Biol. Chem. 272, 17694–1702 (1997).
Gaiano, N., Nye, J.S. & Fishell, G. Radial glial identity is promoted by Notch1 signaling in the murine forebrain. Neuron 26, 395–404 (2000).
Iso, T., Kedes, L. & Hamamori, Y. HES and HERP families: multiple effectors of the Notch signaling pathway. J. Cell. Physiol. 194, 237–255 (2003).
Morita, S., Kojima, T. & Kitamura, T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 7, 1063–1066 (2000).
Tabata, H. & Nakajima, K. Efficient in utero gene transfer system to the developing mouse brain using electroporation: visualization of neuronal migration in the developing cortex. Neuroscience 103, 865–872 (2001).
Acknowledgements
We thank T. Hirano, M. Hibi, T. Taga, K. Nakashima, R. Kageyama, T. Kitamura, T. Iso, T. Sudo, A. Miyajima, T. Kinoshita and R. S. Datta for reagents and advice. We also thank K. Nakajima and H. Tabata for technical help. We are grateful to R. Dolmetsch, M.E. Greenberg, T. Curran, K. Matsumoto and A. Yoshimura for critical reading of the manuscript. We also thank other members of the Gotoh laboratory for their encouragement and helpful discussions. This work is supported by Grants-in-Aid from the Ministry of Education, Science, Sports and Culture of Japan, by PRESTO21 and CREST of the Japan Science and Technology Corporation and VRIA research foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figures
Fig. S1, Fig. S2, Fig. S3 and Fig. S4 (PDF 1534 kb)
Rights and permissions
About this article
Cite this article
Kamakura, S., Oishi, K., Yoshimatsu, T. et al. Hes binding to STAT3 mediates crosstalk between Notch and JAK–STAT signalling. Nat Cell Biol 6, 547–554 (2004). https://doi.org/10.1038/ncb1138
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb1138