Abstract
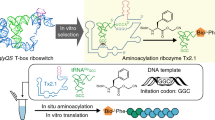
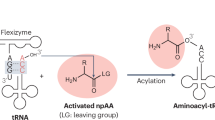
The ability to recognize tRNA identities is essential to the function of the genetic coding system. In translation aminoacyl-tRNA synthetases (ARSs) recognize the identities of tRNAs and charge them with their cognate amino acids. We show that an in vitro–evolved ribozyme can also discriminate between specific tRNAs, and can transfer amino acids to the 3′ ends of cognate tRNAs. The ribozyme interacts with both the CCA-3′ terminus and the anticodon loop of tRNAfMet, and its tRNA specificity is controlled by these interactions. This feature allows us to program the selectivity of the ribozyme toward specific tRNAs, and therefore to tailor effective aminoacyl-transfer catalysts. This method potentially provides a means of generating aminoacyl tRNAs that are charged with non-natural amino acids, which could be incorporated into proteins through cell-free translation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Heckler, T.G. et al. T4 RNA ligase mediated preparation of novel “chemically misacylated” tRNAPhes. Biochemistry 23, 1468–1473 (1984).
Robertson, S.A., Ellman, J.A. & Schultz, P.G. A general and efficient route for chemical aminoacylation of transfer RNAs. J.Am. Chem. Soc. 113, 2711–2729 (1991).
Spirin, A.S. et al. Cell-free translation system capable of producing polypeptides in high yield. Science 242, 1162–1164 (1988).
Kigawa, T. et al. Cell-free production and stable-isotope labeling of milligram quantities of proteins. FEBS Lett. 442, 15–19 (1999).
Liu, D.R., Magliery, T.J., Pastrnak, M. & Schultz, P.G. Engineering a tRNA and aminoacyl-tRNA synthetase for the site-specific incorporation of unnatural amino acids into proteins in vivo. Proc. Natl. Acad. Sci. USA 94, 10092–10097 (1997).
Wang, L., Brock, A., Herberich, B. & Schultz, P.G. Expanding the genetic code of Escherichia coli. Science 292, 498–500 (2001).
Döring, V. et al. Enlarging the amino acid set of Escherichia coli by infiltration of the valine coding pathway. Science 292, 501–504 (2001).
Schimmel, P. & Söll, D. When protein engineering confronts the tRNA world. Proc. Natl. Acad. Sci. USA 94, 10007–10009 (1997).
Liu, D.R. & Schultz, P.G. Progress toward the evolution of an organism with an expanded genetic code. Proc. Natl. Acad. Sci. USA 96, 4780–4785 (1999).
Lohse, P.A. & Szostak, J.W. Ribozyme-catalyzed amino-acid transfer reactions. Nature 381, 442–444 (1996).
Flynn-Charlebois, A., Lee, N. & Suga, H. A single metal ion plays structural and chemical roles in an acyl-transferase ribozyme. Biochemistry 40, 13623–13632 (2001).
Lee, N., Bessho, Y., Wei, K., Szostak, J.W. & Suga, H. Ribozyme-catalyzed tRNA aminoacylation. Nat. Struct. Biol. 7, 28–33 (2000).
Pütz, J. et al. Rapid selection of aminoacyl-tRNAs based on biotinylation of α-NH2 group of charged amino acids. Nucleic Acids Res. 25, 1862–1863 (1997).
Mathews, D.H., Sabina, J., Zuker, M. & Turner, D.H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288, 911–940 (1999).
Arslan, T., Mamaev, S.V., Mamaev, N.V. & Hecht, S.M. Structurally modified firefly luciferase. Effects of amino acid substitution at position 286. J. Am. Chem. Soc. 119, 10877–10887 (1997).
Hohsaka, T., Ashizuka, Y., Murakami, H. & Sisido, M. Incorporation of nonnatural amino acids into streptavidin through in vitro frame-shift suppression. J. Am. Chem. Soc. 118, 9778–9779 (1996).
Acknowledgements
We thank all members of the Suga laboratory for helpful discussions, particularly N. Lee for synthesis of substrates. D.R.W.H. is a grateful recipient of a Royal Society–Fulbright Postdoctoral Fellowship (United Kingdom). This work was supported by US National Institutes of Health and National Science Foundation awards to H.S.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Bessho, Y., Hodgson, D. & Suga, H. A tRNA aminoacylation system for non-natural amino acids based on a programmable ribozyme. Nat Biotechnol 20, 723–728 (2002). https://doi.org/10.1038/nbt0702-723
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nbt0702-723
This article is cited by
-
Cell-free Biosynthesis of Peptidomimetics
Biotechnology and Bioprocess Engineering (2023)
-
Possible Ancestral Functions of the Genetic and RNA Operational Precodes and the Origin of the Genetic System
Origins of Life and Evolution of Biospheres (2021)
-
Ribosome-mediated polymerization of long chain carbon and cyclic amino acids into peptides in vitro
Nature Communications (2020)
-
Incorporation of nonstandard amino acids into proteins: principles and applications
World Journal of Microbiology and Biotechnology (2020)
-
Generation and selection of ribozyme variants with potential application in protein engineering and synthetic biology
Applied Microbiology and Biotechnology (2014)