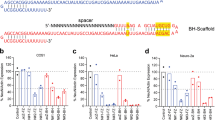
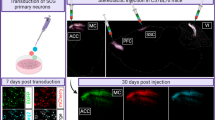
Abstract
Probing gene function in the mammalian brain can be greatly assisted with methods to manipulate the genome of neurons in vivo. The clustered, regularly interspaced, short palindromic repeats (CRISPR)-associated endonuclease (Cas)9 from Streptococcus pyogenes (SpCas9)1 can be used to edit single or multiple genes in replicating eukaryotic cells, resulting in frame-shifting insertion/deletion (indel) mutations and subsequent protein depletion. Here, we delivered SpCas9 and guide RNAs using adeno-associated viral (AAV) vectors to target single (Mecp2) as well as multiple genes (Dnmt1, Dnmt3a and Dnmt3b) in the adult mouse brain in vivo. We characterized the effects of genome modifications in postmitotic neurons using biochemical, genetic, electrophysiological and behavioral readouts. Our results demonstrate that AAV-mediated SpCas9 genome editing can enable reverse genetic studies of gene function in the brain.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Hsu, P.D., Lander, E.S. & Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278 (2014).
Xue, W. et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature 10.1038/nature13589 (6 August 2014).
Yin, H. et al. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat. Biotechnol. 32, 551–553 (2014).
Ding, Q. et al. Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing. Circ. Res. 115, 488–492 (2014).
Wu, Z., Yang, H. & Colosi, P. Effect of genome size on AAV vector packaging. Mol. Ther. 18, 80–86 (2010).
Gray, S.J. et al. Optimizing promoters for recombinant adeno-associated virus-mediated gene expression in the peripheral and central nervous system using self-complementary vectors. Hum. Gene Ther. 22, 1143–1153 (2011).
Ostlund, C. et al. Dynamics and molecular interactions of linker of nucleoskeleton and cytoskeleton (LINC) complex proteins. J. Cell Sci. 122, 4099–4108 (2009).
Chahrour, M. & Zoghbi, H.Y. The story of Rett syndrome: from clinic to neurobiology. Neuron 56, 422–437 (2007).
Chen, R.Z., Akbarian, S., Tudor, M. & Jaenisch, R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat. Genet. 27, 327–331 (2001).
Li, Y. et al. Global transcriptional and translational repression in human-embryonic-stem-cell-derived Rett syndrome neurons. Cell Stem Cell 13, 446–458 (2013).
Zhou, Z. et al. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron 52, 255–269 (2006).
Qi, L.S. et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183 (2013).
Bikard, D. et al. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 41, 7429–7437 (2013).
Gilbert, L.A. et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451 (2013).
Grindberg, R.V. et al. RNA-sequencing from single nuclei. Proc. Natl. Acad. Sci. USA 110, 19802–19807 (2013).
Moretti, P. et al. Learning and memory and synaptic plasticity are impaired in a mouse model of Rett syndrome. J. Neurosci. 26, 319–327 (2006).
Kheirbek, M.A. et al. Differential control of learning and anxiety along the dorsoventral axis of the dentate gyrus. Neuron 77, 955–968 (2013).
Chahrour, M. et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science 320, 1224–1229 (2008).
Hubel, D.H. & Wiesel, T.N. Receptive fields of single neurones in the cat's striate cortex. J. Physiol. (Lond.) 148, 574–591 (1959).
Banerjee, A., Castro, J. & Sur, M. Rett syndrome: genes, synapses, circuits, and therapeutics. Front. Psychiatry 3, 34 (2012).
Chao, H.T., Zoghbi, H.Y. & Rosenmund, C. MeCP2 controls excitatory synaptic strength by regulating glutamatergic synapse number. Neuron 56, 58–65 (2007).
Wood, L., Gray, N.W., Zhou, Z., Greenberg, M.E. & Shepherd, G.M. Synaptic circuit abnormalities of motor-frontal layer 2/3 pyramidal neurons in an RNA interference model of methyl-CpG-binding protein 2 deficiency. J. Neurosci. 29, 12440–12448 (2009).
McGraw, C.M., Samaco, R.C. & Zoghbi, H.Y. Adult neural function requires MeCP2. Science 333, 186 (2011).
Feng, J. et al. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat. Neurosci. 13, 423–430 (2010).
Fu, Y. et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 31, 822–826 (2013).
Hsu, P.D. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832 (2013).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Fonfara, I. et al. Phylogeny of Cas9 determines functional exchangeability of dual-RNA and Cas9 among orthologous type II CRISPR-Cas systems. Nucleic Acids Res. 42, 2577–2590 (2014).
Esvelt, K.M. et al. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat. Methods 10, 1116–1121 (2013).
Platt, R.J. et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455 (2014).
Levitt, N., Briggs, D., Gil, A. & Proudfoot, N.J. Definition of an efficient synthetic poly(A) site. Genes Dev. 3, 1019–1025 (1989).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Sapranauskas, R. et al. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 39, 9275–9282 (2011).
McClure, C., Cole, K.L., Wulff, P., Klugmann, M. & Murray, A.J. Production and titering of recombinant adeno-associated viral vectors. J. Vis. Exp. 57, e3348 (2011).
Konermann, S. et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature 500, 472–476 (2013).
Banker, G. & Goslin, K. Developments in neuronal cell culture. Nature 336, 185–186 (1988).
Swiech, L. et al. CLIP-170 and IQGAP1 cooperatively regulate dendrite morphology. J. Neurosci. 31, 4555–4568 (2011).
Sholl, D.A. Dendritic organization in the neurons of the visual and motor cortices of the cat. J. Anat. 87, 387–406 (1953).
Hagihara, H., Toyama, K., Yamasaki, N. & Miyakawa, T. Dissection of hippocampal dentate gyrus from adult mouse. J. Vis. Exp. 33, 1543 (2009).
Ran, F.A. et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Picelli, S. et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods 10, 1096–1098 (2013).
Fujita, P.A. et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 39, D876–D882 (2011).
Acknowledgements
We thank A. Trevino and C. Le for technical assistance and the entire Zhang lab for technical support and critical discussions; we thank R. Platt (Broad Institute) and H. Worman (Columbia University) for sharing plasmids, R. Rikhye for providing a template for electrophysiology analysis; and X. Yu for statistical discussions. L.S. is a European Molecular Biology Organization (EMBO) Fellow and is supported by the Foundation for Polish Science. M.H. is supported by the Human Frontiers Scientific Program. A.B. holds a postdoctoral fellowship from the Simons Center for the Social Brain. N.H. is an EMBO Fellow and Y.L. is supported by Friends of the McGovern Institute Fellowship. M.S. is supported by grants from the US National Institutes of Health (NIH) (R01EY007023 and R01MH085802) and the Simons Foundation. F.Z. is supported by the National Institute of Mental Health (NIMH) through NIH Director's Pioneer Award (5DP1-MH100706), the NINDS through a NIH Transformative R01 grant (5R01-NS073124), the Keck, Merkin, Vallee, Damon Runyon, Searle Scholars, Klarman Family Foundation, Klingenstein, Poitras and Simons Foundations, and Bob Metcalfe. The authors plan on making the reagents widely available to the academic community through Addgene and to provide software tools via the Zhang lab website (http://www.genome-engineering.org/).
Author information
Authors and Affiliations
Contributions
L.S., M.H. and F.Z. developed the concept and designed experiments. L.S. and M.H. carried out CRISPR-Cas9-related experiments and analyzed data. A.B. designed and performed electrophysiological experiments and analyzed data. N.H., Y.L. and J.T. carried-out RNA sequencing experiments and analyzed data. Y.L. analyzed NGS data. L.S., M.H. and F.Z. wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
F.Z. is a scientific advisor of Editas Medicine and Horizon Discovery. A patent application has been filed relating to this work.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–12 and Supplementary Tables 1–3 (PDF 1449 kb)
Rights and permissions
About this article
Cite this article
Swiech, L., Heidenreich, M., Banerjee, A. et al. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat Biotechnol 33, 102–106 (2015). https://doi.org/10.1038/nbt.3055
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.3055
This article is cited by
-
Acoustically targeted noninvasive gene therapy in large brain volumes
Gene Therapy (2024)
-
Efficient prime editing in mouse brain, liver and heart with dual AAVs
Nature Biotechnology (2024)
-
Removal of a partial genomic duplication restores synaptic transmission and behavior in the MyosinVA mutant mouse Flailer
BMC Biology (2023)
-
Doxycycline-dependent Cas9-expressing pig resources for conditional in vivo gene nullification and activation
Genome Biology (2023)
-
Combinatorial design of nanoparticles for pulmonary mRNA delivery and genome editing
Nature Biotechnology (2023)