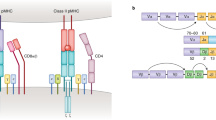
Abstract
Adaptive immune responses often begin with the formation of a molecular complex between a T-cell receptor (TCR) and a peptide antigen bound to a major histocompatibility complex (MHC) molecule. These complexes are highly variable, however, due to the polymorphism of MHC genes, the random, inexact recombination of TCR gene segments, and the vast array of possible self and pathogen peptide antigens. As a result, it has been very difficult to comprehensively study the TCR repertoire or identify and track more than a few antigen-specific T cells in mice or humans. For mouse studies, this had led to a reliance on model antigens and TCR transgenes. The study of limited human clinical samples, in contrast, requires techniques that can simultaneously survey TCR phenotype and function, and TCR reactivity to many T-cell epitopes. Thanks to recent advances in single-cell and cytometry methodologies, as well as high-throughput sequencing of the TCR repertoire, we now have or will soon have the tools needed to comprehensively analyze T-cell responses in health and disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Burnet, F.M. A modification of Jerne's theory of antibody production using the concept of clonal selection. Aust. J. Sci. 20, 67–69 (1957).
Burnet, F.M. The Clonal Selection Theory of Acquired Immunity (Vanderbilt University Press, 1959).
Hulett, H.R., Bonner, W.A., Barrett, J. & Herzenberg, L.A. Cell sorting: automated separation of mammalian cells as a function of intracellular fluorescence. Science 166, 747–749 (1969).
Chattopadhyay, P.K. & Roederer, M. Cytometry: today's technology and tomorrow's horizons. Methods 57, 251–258 (2012).
Han, Q., Bradshaw, E.M., Nilsson, B., Hafler, D.A. & Love, J.C. Multidimensional analysis of the frequencies and rates of cytokine secretion from single cells by quantitative microengraving. Lab Chip 10, 1391–1400 (2010).
Han, Q. et al. Polyfunctional responses by human T cells result from sequential release of cytokines. Proc. Natl. Acad. Sci. USA 109, 1607–1612 (2012).
Varadarajan, N. et al. Rapid, efficient functional characterization and recovery of HIV-specific human CD8+ T cells using microengraving. Proc. Natl. Acad. Sci. USA 109, 3885–3890 (2012).
Betts, M.R. et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood 107, 4781–4789 (2006).
Seder, R.A., Darrah, P.A. & Roederer, M. T-cell quality in memory and protection: implications for vaccine design. Nat. Rev. Immunol. 8, 247–258 (2008).
Makedonas, G. & Betts, M.R. Living in a house of cards: re-evaluating CD8+ T-cell immune correlates against HIV. Immunol. Rev. 239, 109–124 (2011).
Yuan, J. et al. CTLA-4 blockade enhances polyfunctional NY-ESO-1 specific T cell responses in metastatic melanoma patients with clinical benefit. Proc. Natl. Acad. Sci. USA 105, 20410–20415 (2008).
Walker, B.D. & Yu, X.G. Unravelling the mechanisms of durable control of HIV-1. Nat. Rev. Immunol. 13, 487–498 (2013).
Precopio, M.L. et al. Immunization with vaccinia virus induces polyfunctional and phenotypically distinctive CD8+ T cell responses. J. Exp. Med. 204, 1405–1416 (2007).
Yamanaka, Y.J., Gierahn, T.M. & Love, J.C. The dynamic lives of T cells: new approaches and themes. Trends Immunol. 34, 59–66 (2013).
Akram, A. & Inman, R.D. Immunodominance: a pivotal principle in host response to viral infections. Clin. Immunol. 143, 99–115 (2012).
Kiepiela, P. et al. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat. Med. 13, 46–53 (2007).
Pereyra, F. et al. Genetic and immunologic heterogeneity among persons who control HIV infection in the absence of therapy. J. Infect. Dis. 197, 563–571 (2008).
Bowen, D.G. & Walker, C.M. Adaptive immune responses in acute and chronic hepatitis C virus infection. Nature 436, 946–952 (2005).
Hislop, A.D., Annels, N.E., Gudgeon, N.H., Leese, A.M. & Rickinson, A.B. Epitope-specific evolution of human CD8+ T cell responses from primary to persistent phases of Epstein-Barr virus infection. J. Exp. Med. 195, 893–905 (2002).
Newell, E.W. et al. Combinatorial tetramer staining and mass cytometry analysis facilitate T-cell epitope mapping and characterization. Nat. Biotechnol. 31, 623–629 (2013).
Bendall, S.C. et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 332, 687–696 (2011).
Newell, E.W., Sigal, N., Bendall, S.C., Nolan, G.P. & Davis, M.M. Cytometry by time-of-flight shows combinatorial cytokine expression and virus-specific cell niches within a continuum of CD8+ T cell phenotypes. Immunity 36, 142–152 (2012).
Hadrup, S.R. et al. Parallel detection of antigen-specific T-cell responses by multidimensional encoding of MHC multimers. Nat. Methods 6, 520–526 (2009).
Newell, E.W., Klein, L.O., Yu, W. & Davis, M.M. Simultaneous detection of many T-cell specificities using combinatorial tetramer staining. Nat. Methods 6, 497–499 (2009).
Dominguez, M.H. et al. Highly multiplexed quantitation of gene expression on single cells. J. Immunol. Methods 391, 133–145 (2013).
Shalek, A.K. et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature 498, 236–240 (2013).
Warren, E.H., Matsen, F.A.t. & Chou, J. High-throughput sequencing of B- and T-lymphocyte antigen receptors in hematology. Blood 122, 19–22 (2013).
La Gruta, N.L. & Thomas, P.G. Interrogating the relationship between naive and immune antiviral T cell repertoires. Curr. Opin. Virol. 3, 447–451 (2013).
Han, A. et al. Dietary gluten triggers concomitant activation of CD4+ and CD8+ alphabeta T cells and gammadelta T cells in celiac disease. Proc. Natl. Acad. Sci. USA 110, 13073–13078 (2013).
Zhu, J. et al. Immune surveillance by CD8alphaalpha+ skin-resident T cells in human herpes virus infection. Nature 497, 494–497 (2013).
Emerson, R.O. et al. High-throughput sequencing of T cell receptors reveals a homogeneous repertoire of tumor-infiltrating lymphocytes in ovarian cancer. J. Pathol. 231, 433–440 (2013).
Adams, J.J. et al. T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex. Immunity 35, 681–693 (2011).
Birnbaum, M.E., Dong, S. & Garcia, K.C. Diversity-oriented approaches for interrogating T-cell receptor repertoire, ligand recognition, and function. Immunol. Rev. 250, 82–101 (2012).
Davis, M.M. & Bjorkman, P.J. T-cell antigen receptor genes and T-cell recognition. Nature 334, 395–402 (1988).
Aghaeepour, N. et al. Critical assessment of automated flow cytometry data analysis techniques. Nat. Methods 10, 228–238 (2013).
Qiu, P. et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat. Biotechnol. 29, 886–891 (2011).
Finak, G. et al. Mixture models for single-cell assays with applications to vaccine studies. Biostatistics 15, 87–101 (2014).
Amir, E.A. et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol. 31, 545–552 (2013).
Sallusto, F., Lenig, D., Forster, R., Lipp, M. & Lanzavecchia, A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 401, 708–712 (1999).
Romero, P. et al. Four functionally distinct populations of human effector-memory CD8+ T lymphocytes. J. Immunol. 178, 4112–4119 (2007).
Gattinoni, L. et al. A human memory T cell subset with stem cell-like properties. Nat. Med. 17, 1290–1297 (2011).
Kaech, S.M. & Cui, W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat. Rev. Immunol. 12, 749–761 (2012).
Masopust, D. & Schenkel, J.M. The integration of T cell migration, differentiation and function. Nat. Rev. Immunol. 13, 309–320 (2013).
Ornatsky, O., Baranov, V.I., Bandura, D.R., Tanner, S.D. & Dick, J. Multiple cellular antigen detection by ICP-MS. J. Immunol. Methods 308, 68–76 (2006).
Bjornson, Z.B., Nolan, G.P. & Fantl, W.J. Single-cell mass cytometry for analysis of immune system functional states. Curr. Opin. Immunol. 25, 484–494 (2013).
Bodenmiller, B. et al. Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nat. Biotechnol. 30, 858–867 (2012).
Kidd, B.A., Peters, L.A., Schadt, E.E. & Dudley, J.T. Unifying immunology with informatics and multiscale biology. Nat. Immunol. 15, 118–127 (2014).
Shapiro, E., Biezuner, T. & Linnarsson, S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat. Rev. Genet. 14, 618–630 (2013).
Flatz, L. et al. Single-cell gene-expression profiling reveals qualitatively distinct CD8 T cells elicited by different gene-based vaccines. Proc. Natl. Acad. Sci. USA 108, 5724–5729 (2011).
Tang, F. et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 6, 377–382 (2009).
Brunner, K.T., Mauel, J., Cerottini, J.C. & Chapuis, B. Quantitative assay of the lytic action of immune lymphoid cells on 51-Cr-labelled allogeneic target cells in vitro; inhibition by isoantibody and by drugs. Immunology 14, 181–196 (1968).
Peters, P.J. et al. Cytotoxic T lymphocyte granules are secretory lysosomes, containing both perforin and granzymes. J. Exp. Med. 173, 1099–1109 (1991).
Betts, M.R. et al. Sensitive and viable identification of antigen-specific CD8+ T cells by a flow cytometric assay for degranulation. J. Immunol. Methods 281, 65–78 (2003).
Waldrop, S.L., Pitcher, C.J., Peterson, D.M., Maino, V.C. & Picker, L.J. Determination of antigen-specific memory/effector CD4+ T cell frequencies by flow cytometry: evidence for a novel, antigen-specific homeostatic mechanism in HIV-associated immunodeficiency. J. Clin. Invest. 99, 1739–1750 (1997).
De Rosa, S.C. et al. Vaccination in humans generates broad T cell cytokine responses. J. Immunol. 173, 5372–5380 (2004).
Frentsch, M. et al. Direct access to CD4+ T cells specific for defined antigens according to CD154 expression. Nat. Med. 11, 1118–1124 (2005).
Altman, J.D. et al. Phenotypic analysis of antigen-specific T lymphocytes. Science 274, 94–96 (1996).
Davis, M.M., Altman, J.D. & Newell, E.W. Interrogating the repertoire: broadening the scope of peptide-MHC multimer analysis. Nat. Rev. Immunol. 11, 551–558 (2011).
Toebes, M. et al. Design and use of conditional MHC class I ligands. Nat. Med. 12, 246–251 (2006).
Grotenbreg, G.M. et al. Discovery of CD8+ T cell epitopes in Chlamydia trachomatis infection through use of caged class I MHC tetramers. Proc. Natl. Acad. Sci. USA 105, 3831–3836 (2008).
Day, C.L. et al. Ex vivo analysis of human memory CD4 T cells specific for hepatitis C virus using MHC class II tetramers. J. Clin. Invest. 112, 831–842 (2003).
Moon, J.J. et al. Naive CD4+ T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity 27, 203–213 (2007).
Andersen, R.S. et al. Parallel detection of antigen-specific T cell responses by combinatorial encoding of MHC multimers. Nat. Protoc. 7, 891–902 (2012).
Chang, C.X. et al. Sources of diversity in T cell epitope discovery. Front. Biosci. (Landmark Ed.) 16, 3014–3035 (2011).
Assarsson, E. et al. Immunomic analysis of the repertoire of T-cell specificities for influenza A virus in humans. J. Virol. 82, 12241–12251 (2008).
Maciel, M. Jr. et al. Comprehensive analysis of T cell epitope discovery strategies using 17DD yellow fever virus structural proteins and BALB/c (H2d) mice model. Virology 378, 105–117 (2008).
Weiskopf, D. et al. Comprehensive analysis of dengue virus-specific responses supports an HLA-linked protective role for CD8+ T cells. Proc. Natl. Acad. Sci. USA 110, E2046–E2053 (2013).
Hoof, I. et al. Interdisciplinary analysis of HIV-specific CD8+ T cell responses against variant epitopes reveals restricted TCR promiscuity. J. Immunol. 184, 5383–5391 (2010).
Lundegaard, C. et al. NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8–11. Nucleic Acids Res. 36, W509–W512 (2008).
Wee, L.J., Lim, S.J., Ng, L.F. & Tong, J.C. Immunoinformatics: how in silico methods are re-shaping the investigation of peptide immune specificity. Front. Biosci. (Elite Ed.) 4, 311–319 (2012).
Rivino, L. et al. Defining CD8+ T cell determinants during human viral infection in populations of Asian ethnicity. J. Immunol. 191, 4010–4019 (2013).
Yang, J. et al. Multiplex mapping of CD4 T cell epitopes using class II tetramers. Clin. Immunol. 120, 21–32 (2006).
Heemskerk, B., Kvistborg, P. & Schumacher, T.N. The cancer antigenome. EMBO J. 32, 194–203 (2013).
van Rooij, N. et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J. Clin. Oncol. 31, e439–e442 (2013).
Rotzschke, O. et al. Isolation and analysis of naturally processed viral peptides as recognized by cytotoxic T cells. Nature 348, 252–254 (1990).
Marrack, P., Ignatowicz, L., Kappler, J.W., Boymel, J. & Freed, J.H. Comparison of peptides bound to spleen and thymus class II. J. Exp. Med. 178, 2173–2183 (1993).
Fortier, M.H. et al. The MHC class I peptide repertoire is molded by the transcriptome. J. Exp. Med. 205, 595–610 (2008).
Kasuga, K. Comprehensive analysis of MHC ligands in clinical material by immunoaffinity-mass spectrometry. Methods Mol. Biol. 1023, 203–218 (2013).
Baker, E.S. et al. Mass spectrometry for translational proteomics: progress and clinical implications. Genome Med. 4, 63 (2012).
Robins, H.S. et al. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood 114, 4099–4107 (2009).
Robins, H.S. et al. Overlap and effective size of the human CD8+ T cell receptor repertoire. Sci. Transl. Med. 2, 47ra64 (2010).
Venturi, V. et al. A mechanism for TCR sharing between T cell subsets and individuals revealed by pyrosequencing. J. Immunol. 186, 4285–4294 (2011).
Arstila, T.P. et al. A direct estimate of the human alphabeta T cell receptor diversity. Science 286, 958–961 (1999).
Klarenbeek, P.L. et al. Human T-cell memory consists mainly of unexpanded clones. Immunol. Lett. 133, 42–48 (2010).
Dziubianau, M. et al. TCR repertoire analysis by next generation sequencing allows complex differential diagnosis of T cell-related pathology. Am. J. Transplant. 13, 2842–2854 (2013).
Boyd, S.D., Liu, Y., Wang, C., Martin, V. & Dunn-Walters, D.K. Human lymphocyte repertoires in ageing. Curr. Opin. Immunol. 25, 511–515 (2013).
Wu, D. et al. High-throughput sequencing detects minimal residual disease in acute T lymphoblastic leukemia. Sci. Transl. Med. 4, 134ra163 (2012).
DeKosky, B.J. et al. High-throughput sequencing of the paired human immunoglobulin heavy and light chain repertoire. Nat. Biotechnol. 31, 166–169 (2013).
Turchaninova, M.A. et al. Pairing of T-cell receptor chains via emulsion PCR. Eur. J. Immunol. 43, 2507–2515 (2013).
Sollid, L.M. & Jabri, B. Triggers and drivers of autoimmunity: lessons from coeliac disease. Nat. Rev. Immunol. 13, 294–302 (2013).
Reay, P.A., Kantor, R.M. & Davis, M.M. Use of global amino acid replacements to define the requirements for MHC binding and T cell recognition of moth cytochrome c (93–103). J. Immunol. 152, 3946–3957 (1994).
Garcia, K.C., Teyton, L. & Wilson, I.A. Structural basis of T cell recognition. Annu. Rev. Immunol. 17, 369–397 (1999).
Wu, L.C., Tuot, D.S., Lyons, D.S., Garcia, K.C. & Davis, M.M. Two-step binding mechanism for T-cell receptor recognition of peptide MHC. Nature 418, 552–556 (2002).
Janin, J. Protein-protein docking tested in blind predictions: the CAPRI experiment. Mol. Biosyst. 6, 2351–2362 (2010).
Ritchie, D.W. Recent progress and future directions in protein-protein docking. Curr. Protein Pept. Sci. 9, 1–15 (2008).
Reiser, J.B. et al. CDR3 loop flexibility contributes to the degeneracy of TCR recognition. Nat. Immunol. 4, 241–247 (2003).
Su, L.F., Kidd, B.A., Han, A., Kotzin, J.J. & Davis, M.M. Virus-specific CD4+ memory-phenotype T cells are abundant in unexposed adults. Immunity 38, 373–383 (2013).
Su, L.F. & Davis, M.M. Antiviral memory phenotype T cells in unexposed adults. Immunol. Rev. 255, 95–109 (2013).
Parameswaran, P. et al. Convergent antibody signatures in human dengue. Cell Host Microbe 13, 691–700 (2013).
Crawford, F. et al. Use of baculovirus MHC/peptide display libraries to characterize T-cell receptor ligands. Immunol. Rev. 210, 156–170 (2006).
Stadinski, B.D. et al. Chromogranin A is an autoantigen in type 1 diabetes. Nat. Immunol. 11, 225–231 (2010).
Wen, F., Esteban, O. & Zhao, H. Rapid identification of CD4+ T-cell epitopes using yeast displaying pathogen-derived peptide library. J. Immunol. Methods 336, 37–44 (2008).
Harvey, C.J. & Wucherpfennig, K.W. Cracking the code of human T-cell immunity. Nat. Biotechnol. 31, 609–610 (2013).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Newell, E., Davis, M. Beyond model antigens: high-dimensional methods for the analysis of antigen-specific T cells. Nat Biotechnol 32, 149–157 (2014). https://doi.org/10.1038/nbt.2783
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.2783
This article is cited by
-
Yeast display of MHC-II enables rapid identification of peptide ligands from protein antigens (RIPPA)
Cellular & Molecular Immunology (2021)
-
Recent advances in cancer immunotherapy
Discover Oncology (2021)
-
A novel data fusion method for the effective analysis of multiple panels of flow cytometry data
Scientific Reports (2019)
-
BALDR: a computational pipeline for paired heavy and light chain immunoglobulin reconstruction in single-cell RNA-seq data
Genome Medicine (2018)
-
The human skin microbiome
Nature Reviews Microbiology (2018)