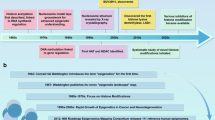
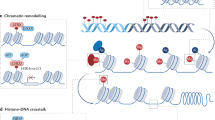
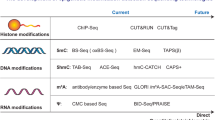
Abstract
Epigenetics is one of the most rapidly expanding fields in biology. The recent characterization of a human DNA methylome at single nucleotide resolution, the discovery of the CpG island shores, the finding of new histone variants and modifications, and the unveiling of genome-wide nucleosome positioning maps highlight the accelerating speed of discovery over the past two years. Increasing interest in epigenetics has been accompanied by technological breakthroughs that now make it possible to undertake large-scale epigenomic studies. These allow the mapping of epigenetic marks, such as DNA methylation, histone modifications and nucleosome positioning, which are critical for regulating gene and noncoding RNA expression. In turn, we are learning how aberrant placement of these epigenetic marks and mutations in the epigenetic machinery is involved in disease. Thus, a comprehensive understanding of epigenetic mechanisms, their interactions and alterations in health and disease, has become a priority in biomedical research.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Esteller, M. Epigenetics in evolution and disease. Lancet 372, S90–S96 (2008).
Waddington, C.H. Introduction to Modern Genetics (Macmillan, 1939).
Rideout, W.M. III, Eggan, K. & Jaenisch, R. Nuclear cloning and epigenetic reprogramming of the genome. Science 293, 1093–1098 (2001).
Fraga, M.F. et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc. Natl. Acad. Sci. USA 102, 10604–10609 (2005).
Kaminsky, Z.A. et al. DNA methylation profiles in monozygotic and dizygotic twins. Nat. Genet. 41, 240–245 (2009).
Javierre, B.M. et al. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 20, 170–179 (2010).
Chi, A.S. & Bernstein, B.E. Developmental biology. Pluripotent chromatin state. Science 323, 220–221 (2009).
Meissner, A. et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454, 766–770 (2008).
Meissner, A. Epigenetic modifications in pluripotent and differentiated cells. Nat. Biotechnol. 28, 1079–1088 (2010).
Esteller, M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene 21, 5427–5440 (2002).
Esteller, M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet. 8, 286–298 (2007).
Straussman, R. et al. Developmental programming of CpG island methylation profiles in the human genome. Nat. Struct. Mol. Biol. 16, 564–571 (2009).
Kacem, S. & Feil, R. Chromatin mechanisms in genomic imprinting. Mamm. Genome 20, 544–556 (2009).
Reik, W. & Lewis, A. Co-evolution of X-chromosome inactivation and imprinting in mammals. Nat. Rev. Genet. 6, 403–410 (2005).
Esteller, M. Epigenetic gene silencing in cancer: the DNA hypermethylome. Hum. Mol. Genet. 16 Spec No 1, R50–R59 (2007).
Lopez-Serra, L. & Esteller, M. Proteins that bind methylated DNA and human cancer: reading the wrong words. Br. J. Cancer 98, 1881–1885 (2008).
Kuroda, A. et al. Insulin gene expression is regulated by DNA methylation. PLoS ONE 4, e6953 (2009).
Thomson, J.P. et al. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature 464, 1082–1086 (2010).
Irizarry, R.A. et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat. Genet. 41, 178–186 (2009).
Doi, A. et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat. Genet. 41, 1350–1353 (2009).
Ji, H. et al. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature 467, 338–342 (2010).
Hellman, A. & Chess, A. Gene body-specific methylation on the active X chromosome. Science 315, 1141–1143 (2007).
Zilberman, D., Gehring, M., Tran, R.K., Ballinger, T. & Henikoff, S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 39, 61–69 (2007).
Zhao, Z. et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat. Genet. 38, 1341–1347 (2006).
Espada, J. et al. Epigenetic disruption of ribosomal RNA genes and nucleolar architecture in DNA methyltransferase 1 (Dnmt1) deficient cells. Nucleic Acids Res. 35, 2191–2198 (2007).
Horike, S., Cai, S., Miyano, M., Cheng, J.F. & Kohwi-Shigematsu, T. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat. Genet. 37, 31–40 (2005).
van Steensel, B. & Dekker, J. Genomics tools for unraveling chromosome architecture. Nat. Biotechnol. 28, 1089–1095 (2010).
Lister, R. et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462, 315–322 (2009).
Laurent, L. et al. Dynamic changes in the human methylome during differentiation. Genome Res. 20, 320–331 (2010).
Kriaucionis, S. & Heintz, N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324, 929–930 (2009).
Berman, B.P., Weisenberger, D.J. & Laird, P.W. Locking in on the human methylome. Nat. Biotechnol. 27, 341–342 (2009).
Weisenberger, D.J. et al. DNA methylation analysis by digital bisulfite genomic sequencing and digital MethyLight. Nucleic Acids Res. 36, 4689–4698 (2008).
Down, T.A. et al. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat. Biotechnol. 26, 779–785 (2008).
Laird, P.W. Principles and challenges of genome-wide DNA methylation analysis. Nat. Rev. Genet. 11, 191–203 (2010).
Bourc'his, D., Xu, G.L., Lin, C.S., Bollman, B. & Bestor, T.H. Dnmt3L and the establishment of maternal genomic imprints. Science 294, 2536–2539 (2001).
Chen, Z.X., Mann, J.R., Hsieh, C.L., Riggs, A.D. & Chedin, F. Physical and functional interactions between the human DNMT3L protein and members of the de novo methyltransferase family. J. Cell. Biochem. 95, 902–917 (2005).
Holz-Schietinger, C. & Reich, N.O. The inherent processivity of the human de novo DNA methyltransferase 3A (DNMT3A) is enhanced by DNMT3L. J. Biol. Chem. 285, 29091–29100 (2010).
Chuang, L.S. et al. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science 277, 1996–2000 (1997).
Bostick, M. et al. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317, 1760–1764 (2007).
Jones, P.A. & Liang, G. Rethinking how DNA methylation patterns are maintained. Nat. Rev. Genet. 10, 805–811 (2009).
Jeong, S. et al. Selective anchoring of DNA methyltransferases 3A and 3B to nucleosomes containing methylated DNA. Mol. Cell. Biol. 29, 5366–5376 (2009).
Goll, M.G. et al. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 311, 395–398 (2006).
Ooi, S.K. et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448, 714–717 (2007).
Tachibana, M., Matsumura, Y., Fukuda, M., Kimura, H. & Shinkai, Y. G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription. EMBO J. 27, 2681–2690 (2008).
Zhao, Q. et al. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat. Struct. Mol. Biol. 16, 304–311 (2009).
Esteve, P.O. et al. Regulation of DNMT1 stability through SET7-mediated lysine methylation in mammalian cells. Proc. Natl. Acad. Sci. USA 106, 5076–5081 (2009).
Wang, J. et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat. Genet. 41, 125–129 (2009).
Mosher, R.A. & Melnyk, C.W. siRNAs and DNA methylation: seedy epigenetics. Trends Plant Sci. 15, 204–210 (2010).
Matzke, M.A. & Birchler, J.A. RNAi-mediated pathways in the nucleus. Nat. Rev. Genet. 6, 24–35 (2005).
Vrbsky, J. et al. siRNA-mediated methylation of Arabidopsis telomeres. PLoS Genet. 6, e1000986 (2010).
Zaratiegui, M., Irvine, D.V. & Martienssen, R.A. Noncoding RNAs and gene silencing. Cell 128, 763–776 (2007).
Kouzarides, T. Chromatin modifications and their function. Cell 128, 693–705 (2007).
Daujat, S., Zeissler, U., Waldmann, T., Happel, N. & Schneider, R. HP1 binds specifically to Lys26-methylated histone H1.4, whereas simultaneous Ser27 phosphorylation blocks HP1 binding. J. Biol. Chem. 280, 38090–38095 (2005).
Rando, O.J. & Chang, H.Y. Genome-wide views of chromatin structure. Annu. Rev. Biochem. 78, 245–271 (2009).
Huertas, D., Sendra, R. & Munoz, P. Chromatin dynamics coupled to DNA repair. Epigenetics 4, 31–42 (2009).
Luco, R.F. et al. Regulation of alternative splicing by histone modifications. Science 327, 996–1000 (2010).
Li, B., Carey, M. & Workman, J.L. The role of chromatin during transcription. Cell 128, 707–719 (2007).
Karlic, R., Chung, H.R., Lasserre, J., Vlahovicek, K. & Vingron, M. Histone modification levels are predictive for gene expression. Proc. Natl. Acad. Sci. USA 107, 2926–2931 (2010).
Moazed, D. Small RNAs in transcriptional gene silencing and genome defence. Nature 457, 413–420 (2009).
Grewal, S.I. & Jia, S. Heterochromatin revisited. Nat. Rev. Genet. 8, 35–46 (2007).
Khalil, A.M. et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA 106, 11667–11672 (2009).
Chow, J. & Heard, E. X inactivation and the complexities of silencing a sex chromosome. Curr. Opin. Cell Biol. 21, 359–366 (2009).
Agrelo, R. & Wutz, A. X inactivation and disease. Semin. Cell Dev. Biol. 21, 194–200 (2010).
Rinn, J.L. et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323 (2007).
Santos-Rosa, H. et al. Histone H3 tail clipping regulates gene expression. Nat. Struct. Mol. Biol. 16, 17–22 (2009).
Wang, Z. et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 40, 897–903 (2008).
Duan, Q., Chen, H., Costa, M. & Dai, W. Phosphorylation of H3S10 blocks the access of H3K9 by specific antibodies and histone methyltransferase. Implication in regulating chromatin dynamics and epigenetic inheritance during mitosis. J. Biol. Chem. 283, 33585–33590 (2008).
Nakanishi, S. et al. Histone H2BK123 monoubiquitination is the critical determinant for H3K4 and H3K79 trimethylation by COMPASS and Dot1. J. Cell Biol. 186, 371–377 (2009).
Ernst, J. & Kellis, M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat. Biotechnol. 28, 817–825 (2010).
Mikkelsen, T.S. et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553–560 (2007).
Bernstein, B.E. et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 (2006).
Fuks, F. et al. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J. Biol. Chem. 278, 4035–4040 (2003).
Bhaumik, S.R., Smith, E. & Shilatifard, A. Covalent modifications of histones during development and disease pathogenesis. Nat. Struct. Mol. Biol. 14, 1008–1016 (2007).
Chang, B., Chen, Y., Zhao, Y. & Bruick, R.K. JMJD6 is a histone arginine demethylase. Science 318, 444–447 (2007).
Chi, P., Allis, C.D. & Wang, G.G. Covalent histone modifications–miswritten, misinterpreted and mis-erased in human cancers. Nat. Rev. Cancer 10, 457–469 (2010).
Wang, Z. et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138, 1019–1031 (2009).
Schones, D.E. et al. Dynamic regulation of nucleosome positioning in the human genome. Cell 132, 887–898 (2008).
Cairns, B.R. The logic of chromatin architecture and remodelling at promoters. Nature 461, 193–198 (2009).
Chodavarapu, R.K. et al. Relationship between nucleosome positioning and DNA methylation. Nature 466, 388–392 (2010).
Getun, I.V., Wu, Z.K., Khalil, A.M. & Bois, P.R. Nucleosome occupancy landscape and dynamics at mouse recombination hotspots. EMBO Rep. 11, 555–560 (2010).
Zilberman, D., Coleman-Derr, D., Ballinger, T. & Henikoff, S. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature 456, 125–129 (2008).
Harikrishnan, K.N. et al. Brahma links the SWI/SNF chromatin-remodeling complex with MeCP2-dependent transcriptional silencing. Nat. Genet. 37, 254–264 (2005).
Wysocka, J. et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442, 86–90 (2006).
Lal, A. et al. miR-24-mediated downregulation of H2AX suppresses DNA repair in terminally differentiated blood cells. Nat. Struct. Mol. Biol. 16, 492–498 (2009).
Yoo, A.S., Staahl, B.T., Chen, L. & Crabtree, G.R. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature 460, 642–646 (2009).
Ho, L. & Crabtree, G.R. Chromatin remodelling during development. Nature 463, 474–484 (2010).
Reisman, D., Glaros, S. & Thompson, E.A. The SWI/SNF complex and cancer. Oncogene 28, 1653–1668 (2009).
Clapier, C.R. & Cairns, B.R. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78, 273–304 (2009).
Sharma, S., Kelly, T.K. & Jones, P.A. Epigenetics in cancer. Carcinogenesis 31, 27–36 (2009).
Goelz, S.E., Vogelstein, B., Hamilton, S.R. & Feinberg, A.P. Hypomethylation of DNA from benign and malignant human colon neoplasms. Science 228, 187–190 (1985).
Gaudet, F. et al. Induction of tumors in mice by genomic hypomethylation. Science 300, 489–492 (2003).
Wilson, A.S., Power, B.E. & Molloy, P.L. DNA hypomethylation and human diseases. Biochim. Biophys. Acta 1775, 138–162 (2007).
Futscher, B.W. et al. Aberrant methylation of the maspin promoter is an early event in human breast cancer. Neoplasia 6, 380–389 (2004).
Futscher, B.W. et al. Role for DNA methylation in the control of cell type specific maspin expression. Nat. Genet. 31, 175–179 (2002).
Bettstetter, M. et al. Elevated nuclear maspin expression is associated with microsatellite instability and high tumour grade in colorectal cancer. J. Pathol. 205, 606–614 (2005).
Ito, Y. et al. Somatically acquired hypomethylation of IGF2 in breast and colorectal cancer. Hum. Mol. Genet. 17, 2633–2643 (2008).
Li, M. et al. Sensitive digital quantification of DNA methylation in clinical samples. Nat. Biotechnol. 27, 858–863 (2009).
Kelly, T.K., De Carvalho, D.D. & Peter A Jones, P.A. Epigenetic modifications as therapeutic targets. Nat. Biotechnol. 28, 1069–1078 (2010).
Melo, S.A. et al. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat. Genet. 41, 365–370 (2009).
Saito, Y. et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell 9, 435–443 (2006).
Lujambio, A. et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 67, 1424–1429 (2007).
Lujambio, A. et al. A microRNA DNA methylation signature for human cancer metastasis. Proc. Natl. Acad. Sci. USA 105, 13556–13561 (2008).
Di Croce, L. et al. Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science 295, 1079–1082 (2002).
Frigola, J. et al. Epigenetic remodeling in colorectal cancer results in coordinate gene suppression across an entire chromosome band. Nat. Genet. 38, 540–549 (2006).
Miremadi, A., Oestergaard, M.Z., Pharoah, P.D. & Caldas, C. Cancer genetics of epigenetic genes. Hum. Mol. Genet. 16 Spec No 1, R28–R49 (2007).
Garzon, R. et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood 113, 6411–6418 (2009).
Fraga, M.F. et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat. Genet. 37, 391–400 (2005).
Zhu, P. et al. Induction of HDAC2 expression upon loss of APC in colorectal tumorigenesis. Cancer Cell 5, 455–463 (2004).
Ropero, S. et al. A truncating mutation of HDAC2 in human cancers confers resistance to histone deacetylase inhibition. Nat. Genet. 38, 566–569 (2006).
Vaquero, A., Sternglanz, R. & Reinberg, D. NAD+-dependent deacetylation of H4 lysine 16 by class III HDACs. Oncogene 26, 5505–5520 (2007).
Noonan, E.J. et al. miR-449a targets HDAC-1 and induces growth arrest in prostate cancer. Oncogene 28, 1714–1724 (2009).
Moore, S.D. et al. Uterine leiomyomata with t(10;17) disrupt the histone acetyltransferase MORF. Cancer Res. 64, 5570–5577 (2004).
Bryan, E.J. et al. Mutation analysis of EP300 in colon, breast and ovarian carcinomas. Int. J. Cancer 102, 137–141 (2002).
Hamamoto, R. et al. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat. Cell Biol. 6, 731–740 (2004).
Kondo, Y. et al. Alterations of DNA methylation and histone modifications contribute to gene silencing in hepatocellular carcinomas. Hepatol. Res. 37, 974–983 (2007).
Vire, E. et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature 439, 871–874 (2006).
Dalgliesh, G.L. et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature 463, 360–363 (2010).
Gupta, R.A. et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464, 1071–1076 (2010).
Varambally, S. et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science 322, 1695–1699 (2008).
Berdasco, M. et al. Epigenetic inactivation of the Sotos overgrowth syndrome gene histone methyltransferase NSD1 in human neuroblastoma and glioma. Proc. Natl. Acad. Sci. USA 106, 21830–21835 (2009).
Jones, B. et al. The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure. PLoS Genet. 4, e1000190 (2008).
Jacinto, F.V., Ballestar, E. & Esteller, M. Impaired recruitment of the histone methyltransferase DOT1L contributes to the incomplete reactivation of tumor suppressor genes upon DNA demethylation. Oncogene 28, 4212–4224 (2009).
Krivtsov, A.V. et al. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell 14, 355–368 (2008).
Wang, P. et al. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol. Cell. Biol. 29, 6074–6085 (2009).
Shi, Y. Histone lysine demethylases: emerging roles in development, physiology and disease. Nat. Rev. Genet. 8, 829–833 (2007).
Dawson, M.A. et al. JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin. Nature 461, 819–822 (2009).
Medina, P.P. & Sanchez-Cespedes, M. Involvement of the chromatin-remodeling factor BRG1/SMARCA4 in human cancer. Epigenetics 3, 64–68 (2008).
Naidu, S.R., Love, I.M., Imbalzano, A.N., Grossman, S.R. & Androphy, E.J. The SWI/SNF chromatin remodeling subunit BRG1 is a critical regulator of p53 necessary for proliferation of malignant cells. Oncogene 28, 2492–2501 (2009).
Roberts, C.W. & Orkin, S.H. The SWI/SNF complex–chromatin and cancer. Nat. Rev. Cancer 4, 133–142 (2004).
Lin, J.C. et al. Role of nucleosomal occupancy in the epigenetic silencing of the MLH1 CpG island. Cancer Cell 12, 432–444 (2007).
Mulero-Navarro, S. & Esteller, M. Chromatin remodeling factor CHD5 is silenced by promoter CpG island hypermethylation in human cancer. Epigenetics 3, 210–215 (2008).
Sporn, J.C. et al. Histone macroH2A isoforms predict the risk of lung cancer recurrence. Oncogene 28, 3423–3428 (2009).
Gibbs, J.R. et al. Abundant quantitative trait Loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 6, e1000952 (2010).
Wynder, C., Hakimi, M.A., Epstein, J.A., Shilatifard, A. & Shiekhattar, R. Recruitment of MLL by HMG-domain protein iBRAF promotes neural differentiation. Nat. Cell Biol. 7, 1113–1117 (2005).
Urdinguio, R.G., Sanchez-Mut, J.V. & Esteller, M. Epigenetic mechanisms in neurological diseases: genes, syndromes, and therapies. Lancet Neurol. 8, 1056–1072 (2009).
Hite, K.C., Adams, V.H. & Hansen, J.C. Recent advances in MeCP2 structure and function. Biochem. Cell Biol. 87, 219–227 (2009).
Chahrour, M. et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science 320, 1224–1229 (2008).
Urdinguio, R.G. et al. Mecp2-null mice provide new neuronal targets for Rett syndrome. PLoS ONE 3, e3669 (2008).
Nomura, T. et al. MeCP2-dependent repression of an imprinted miR-184 released by depolarization. Hum. Mol. Genet. 17, 1192–1199 (2008).
Urdinguio, R.G. et al. Disrupted microRNA expression caused by Mecp2 loss in a mouse model of Rett syndrome. Epigenetics 5, 656–663 (2010).
Alarcon, J.M. et al. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron 42, 947–959 (2004).
Clayton, A.L., Rose, S., Barratt, M.J. & Mahadevan, L.C. Phosphoacetylation of histone H3 on c-fos- and c-jun-associated nucleosomes upon gene activation. EMBO J. 19, 3714–3726 (2000).
De Sario, A. Clinical and molecular overview of inherited disorders resulting from epigenomic dysregulation. Eur. J. Med. Genet. 52, 363–372 (2009).
Gheldof, N., Tabuchi, T.M. & Dekker, J. The active FMR1 promoter is associated with a large domain of altered chromatin conformation with embedded local histone modifications. Proc. Natl. Acad. Sci. USA 103, 12463–12468 (2006).
Pieper, H.C. et al. Different methylation of the TNF-alpha promoter in cortex and substantia nigra: Implications for selective neuronal vulnerability. Neurobiol. Dis. 32, 521–527 (2008).
Murgatroyd, C. et al. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat. Neurosci. 12, 1559–1566 (2009).
Robertson, K.D. DNA methylation and human disease. Nat. Rev. Genet. 6, 597–610 (2005).
Herman, D. et al. Histone deacetylase inhibitors reverse gene silencing in Friedreich's ataxia. Nat. Chem. Biol. 2, 551–558 (2006).
Al-Mahdawi, S. et al. The Friedreich ataxia GAA repeat expansion mutation induces comparable epigenetic changes in human and transgenic mouse brain and heart tissues. Hum. Mol. Genet. 17, 735–746 (2008).
Kleine-Kohlbrecher, D. et al. A functional link between the histone demethylase PHF8 and the transcription factor ZNF711 in X-linked mental retardation. Mol. Cell 38, 165–178 (2010).
Kumari, D. & Usdin, K. Chromatin remodeling in the noncoding repeat expansion diseases. J. Biol. Chem. 284, 7413–7417 (2009).
Jin, B. et al. DNA methyltransferase 3B (DNMT3B) mutations in ICF syndrome lead to altered epigenetic modifications and aberrant expression of genes regulating development, neurogenesis and immune function. Hum. Mol. Genet. 17, 690–709 (2008).
Pan, W. et al. MicroRNA-21 and microRNA-148a contribute to DNA hypomethylation in lupus CD4+ T cells by directly and indirectly targeting DNA methyltransferase 1. J. Immunol. 184, 6773–6781 (2010).
Javierre, B.M., Esteller, M. & Ballestar, E. Epigenetic connections between autoimmune disorders and haematological malignancies. Trends Immunol. 29, 616–623 (2008).
Karouzakis, E., Gay, R.E., Gay, S. & Neidhart, M. Epigenetic control in rheumatoid arthritis synovial fibroblasts. Nat. Rev. Rheumatol. 5, 266–272 (2009).
Mishra, N., Brown, D.R., Olorenshaw, I.M. & Kammer, G.M. Trichostatin A reverses skewed expression of CD154, interleukin-10, and interferon-gamma gene and protein expression in lupus T cells. Proc. Natl. Acad. Sci. USA 98, 2628–2633 (2001).
Vanden Berghe, W. et al. Keeping up NF-kappaB appearances: epigenetic control of immunity or inflammation-triggered epigenetics. Biochem. Pharmacol. 72, 1114–1131 (2006).
Huber, L.C., Stanczyk, J., Jungel, A. & Gay, S. Epigenetics in inflammatory rheumatic diseases. Arthritis Rheum. 56, 3523–3531 (2007).
Miao, F. et al. Lymphocytes from patients with type 1 diabetes display a distinct profile of chromatin histone H3 lysine 9 dimethylation: an epigenetic study in diabetes. Diabetes 57, 3189–3198 (2008).
Ajiro, K. Histone H2B phosphorylation in mammalian apoptotic cells. An association with DNA fragmentation. J. Biol. Chem. 275, 439–443 (2000).
Hurd, P.J. et al. Phosphorylation of histone H3 Thr-45 is linked to apoptosis. J. Biol. Chem. 284, 16575–16583 (2009).
van Bavel, C.C. et al. Apoptosis-induced histone H3 methylation is targeted by autoantibodies in systemic lupus erythematosus. Ann. Rheum. Dis. published online doi: 10.1136/ard.2010.129320 (10 August 2010).
Dieker, J.W. et al. Apoptosis-induced acetylation of histones is pathogenic in systemic lupus erythematosus. Arthritis Rheum. 56, 1921–1933 (2007).
Van Bavel, J.J. & Cunningham, W.A. Self-categorization with a novel mixed-race group moderates automatic social and racial biases. Pers. Soc. Psychol. Bull. 35, 321–335 (2009).
Verlaan, D.J. et al. Allele-specific chromatin remodeling in the ZPBP2/GSDMB/ORMDL3 locus associated with the risk of asthma and autoimmune disease. Am. J. Hum. Genet. 85, 377–393 (2009).
Turunen, M.P., Aavik, E. & Yla-Herttuala, S. Epigenetics and atherosclerosis. Biochim. Biophys. Acta 1790, 886–891 (2009).
Movassagh, M. et al. Differential DNA methylation correlates with differential expression of angiogenic factors in human heart failure. PLoS ONE 5, e8564 (2010).
Hang, C.T. et al. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature 466, 62–67 (2010).
Symonds, M.E., Sebert, S.P., Hyatt, M.A. & Budge, H. Nutritional programming of the metabolic syndrome. Nat. Rev. Endocrinol. 5, 604–610 (2009).
Zeng, W. et al. Specific loss of histone H3 lysine 9 trimethylation and HP1gamma/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD). PLoS Genet. 5, e1000559 (2009).
Wilkins-Haug, L. Epigenetics and assisted reproduction. Curr. Opin. Obstet. Gynecol. 21, 201–206 (2009).
Tahiliani, M. et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 (2009).
Ito, S. et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466, 1129–1133 (2010).
Ghildiyal, M. & Zamore, P.D. Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 10, 94–108 (2009).
Mattick, J.S. The genetic signatures of noncoding RNAs. PLoS Genet. 5, e1000459 (2009).
Bernstein, B. The NIH Roadmap Epigenome Mapping Consortium. Nat. Biotechnol. 28, 1045–1048 (2010).
Satterlee, J. Tackling the epigenome: challenges and opportunities for collaborative efforts. Nat. Biotechnol. 28, 1039–1044 (2010).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Portela, A., Esteller, M. Epigenetic modifications and human disease. Nat Biotechnol 28, 1057–1068 (2010). https://doi.org/10.1038/nbt.1685
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.1685