Abstract
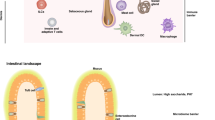
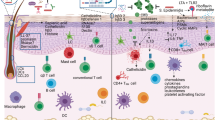
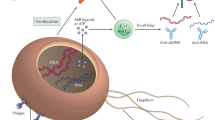
The skin is a complex and dynamic ecosystem that is inhabited by bacteria, archaea, fungi and viruses. These microbes—collectively referred to as the skin microbiota—are fundamental to skin physiology and immunity. Interactions between skin microbes and the host can fall anywhere along the continuum between mutualism and pathogenicity. In this Review, we highlight how host–microbe interactions depend heavily on context, including the state of immune activation, host genetic predisposition, barrier status, microbe localization, and microbe–microbe interactions. We focus on how context shapes the complex dialogue between skin microbes and the host, and the consequences of this dialogue for health and disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
25 January 2018
In the HTML version of this Review, the two corresponding authors were incorrectly listed as Michael A. Fischbach and Y. Erin Chen, instead of Michael A. Fischbach and Yasmine Belkaid.
References
De Luca, C. & Valacchi, G. Surface lipids as multifunctional mediators of skin responses to environmental stimuli. Mediators Inflamm. 2010, 321494 (2010)
van Smeden, J. & Bouwstra, J. A. Stratum corneum lipids: their role for the skin barrier function in healthy subjects and atopic dermatitis patients. Curr. Probl. Dermatol. 49, 8–26 (2016)
Niyonsaba, F., Kiatsurayanon, C., Chieosilapatham, P. & Ogawa, H. Friends or foes? Host defense (antimicrobial) peptides and proteins in human skin diseases. Exp. Dermatol. 26, 989–998 (2017)
Bek-Thomsen, M., Lomholt, H. B., Scavenius, C., Enghild, J. J. & Brüggemann, H. Proteome analysis of human sebaceous follicle infundibula extracted from healthy and acne-affected skin. PLoS One 9, e107908 (2014)
Lee, D.-Y. et al. Sebocytes express functional cathelicidin antimicrobial peptides and can act to kill propionibacterium acnes. J. Invest. Dermatol. 128, 1863–1866 (2008)
Boncheva, M. The physical chemistry of the stratum corneum lipids. Int. J. Cosmet. Sci. 36, 505–515 (2014)
Matard, B. et al. First evidence of bacterial biofilms in the anaerobe part of scalp hair follicles: a pilot comparative study in folliculitis decalvans. J. Eur. Acad. Dermatol. Venereol. 27, 853–860 (2013)
Puhvel, S. M., Reisner, R. M. & Amirian, D. A. Quantification of bacteria in isolated pilosebaceous follicles in normal skin. J. Invest. Dermatol. 65, 525–531 (1975)
Donaldson, G. P., Lee, S. M. & Mazmanian, S. K. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 14, 20–32 (2016)
Peterson, L. W. & Artis, D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat. Rev. Immunol. 14, 141–153 (2014)
Albenberg, L. et al. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology 147, 1055–1063.e8 (2014)
Lind Due, V., Bonde, J., Kann, T. & Perner, A. Extremely low oxygen tension in the rectal lumen of human subjects. Acta Anaesthesiol. Scand. 47, 372 (2003)
Crompton, D. W. T., Shrimpton, D. H. & Silver, I. A. Measurements of the oxygen tension in the lumen of the small intestine of the domestic duck. J. Exp. Biol. 43, 473–478 (1965)
Strauss, J. S., Pochi, P. E. & Downing, D. T. The sebaceous glands: twenty-five years of progress. J. Invest. Dermatol. 67, 90–97 (1976)
Nicolaides, N. Skin lipids: their biochemical uniqueness. Science 186, 19–26 (1974)
Drake, D. R., Brogden, K. A., Dawson, D. V. & Wertz, P. W. Thematic review series: skin lipids. Antimicrobial lipids at the skin surface. J. Lipid Res. 49, 4–11 (2008)
Puhvel, S. M ., Reisner, R. M. & Sakamoto, M. Analysis of lipid composition of isolated human sebaceous gland homogenates after incubation with cutaneous bacteria. Thin-layer chromatography. J. Invest. Dermatol. 64, 406–411 (1975). This study showed that common skin bacteria, such as Cutibacterium (Propionibacterium ) species and Staphylococcus epidermidis , can modify skin lipids through hydrolysis of triglycerides and esterification of cholesterol, and that these enzymatic activities can be modified by other skin features, such as pH.
Sanford, J. A. et al. Inhibition of HDAC8 and HDAC9 by microbial short-chain fatty acids breaks immune tolerance of the epidermis to TLR ligands. Sci. Immunol. 1, eaah4609 (2016)
Scholz, C. F. P. & Kilian, M. The natural history of cutaneous propionibacteria, and reclassification of selected species within the genus Propionibacterium to the proposed novel genera Acidipropionibacterium gen. nov., Cutibacterium gen. nov. and Pseudopropionibacterium gen. nov. Int. J. Syst. Evol. Microbiol. 66, 4422–4432 (2016)
Grice, E. A . et al. Topographical and temporal diversity of the human skin microbiome. Science 324, 1190–1192 (2009). Using 16S ribosomal RNA sequencing, this study provided a metagenomic analysis of the human skin microbiome and described previously unappreciated bacterial diversity at different skin sites.
Oh, J. et al. Biogeography and individuality shape function in the human skin metagenome. Nature 514, 59–64 (2014)
Byrd, A. L. et al. Staphylococcus aureus and Staphylococcus epidermidis strain diversity underlying pediatric atopic dermatitis. Sci. Transl. Med. 9, eaal4651 (2017). This study highlights the utility of shotgun metagenomic sequencing over 16S ribosomal RNA sequencing to assess how strain differences within the same Staphylococcus epidermidis species can contribute to disease.
Chu, D. M. et al. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat. Med. 23, 314–326 (2017)
Oh, J., Conlan, S., Polley, E. C., Segre, J. A. & Kong, H. H. Shifts in human skin and nares microbiota of healthy children and adults. Genome Med. 4, 77 (2012)
Oh, J., Byrd, A. L., Park, M., Kong, H. H. & Segre, J. A. Temporal stability of the human skin microbiome. Cell 165, 854–866 (2016)
Kong, H. H . et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 22, 850–859 (2012). This study was one of the first to use metagenomic sequencing to characterize dysbiosis in inflammatory skin diseases, showing that atopic dermatitis flares are associated not only with blooms of Staphylococcus aureus but also with significant decreases in overall skin microbial diversity.
Otto, M. Staphylococcus epidermidis—the ‘accidental’ pathogen. Nat. Rev. Microbiol. 7, 555–567 (2009)
Ramsey, M. M., Freire, M. O., Gabrilska, R. A., Rumbaugh, K. P. & Lemon, K. P. Staphylococcus aureus shifts toward commensalism in response to Corynebacterium species. Front. Microbiol. 7, 1230 (2016)
Naik, S . et al. Compartmentalized control of skin immunity by resident commensals. Science 337, 1115–1119 (2012). This study demonstrated that skin-resident commensal bacteria are critical for establishing skin immune homeostasis and that this process occurs through an intact, uninflamed skin barrier.
Cebra, J. J. Influences of microbiota on intestinal immune system development. Am. J. Clin. Nutr. 69, 1046S–1051S (1999)
Chehoud, C. et al. Complement modulates the cutaneous microbiome and inflammatory milieu. Proc. Natl Acad. Sci. USA 110, 15061–15066 (2013)
Nagy, I. et al. Propionibacterium acnes and lipopolysaccharide induce the expression of antimicrobial peptides and proinflammatory cytokines/chemokines in human sebocytes. Microbes Infect. 8, 2195–2205 (2006)
Christensen, G. J. M. et al. Antagonism between Staphylococcus epidermidis and Propionibacterium acnes and its genomic basis. BMC Genomics 17, 152 (2016)
Cogen, A. L. et al. Selective antimicrobial action is provided by phenol-soluble modulins derived from Staphylococcus epidermidis, a normal resident of the skin. J. Invest. Dermatol. 130, 192–200 (2010)
Nakatsuji, T. et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci. Transl. Med. 9, eaah4680 (2017)
Källenius, G., Correia-Neves, M., Buteme, H., Hamasur, B. & Svenson, S. B. Lipoarabinomannan, and its related glycolipids, induce divergent and opposing immune responses to Mycobacterium tuberculosis depending on structural diversity and experimental variations. Tuberculosis (Edinb.) 96, 120–130 (2016)
Afonso-Barroso, A. et al. Lipoarabinomannan mannose caps do not affect mycobacterial virulence or the induction of protective immunity in experimental animal models of infection and have minimal impact on in vitro inflammatory responses. Cell. Microbiol. 15, 660–674 (2013)
Briken, V., Porcelli, S. A., Besra, G. S. & Kremer, L. Mycobacterial lipoarabinomannan and related lipoglycans: from biogenesis to modulation of the immune response. Mol. Microbiol. 53, 391–403 (2004)
Chatterjee, D. & Khoo, K.-H. Mycobacterial lipoarabinomannan: an extraordinary lipoheteroglycan with profound physiological effects. Glycobiology 8, 113–120 (1998)
Dao, D. N. et al. Mycobacterium tuberculosis lipomannan induces apoptosis and interleukin-12 production in macrophages. Infect. Immun. 72, 2067–2074 (2004)
Doz, E. et al. Acylation determines the toll-like receptor (TLR)-dependent positive versus TLR2-, mannose receptor-, and SIGNR1-independent negative regulation of pro-inflammatory cytokines by mycobacterial lipomannan. J. Biol. Chem. 282, 26014–26025 (2007)
Fukuda, T. et al. Critical roles for lipomannan and lipoarabinomannan in cell wall integrity of mycobacteria and pathogenesis of tuberculosis. MBio 4, e00472–e12 (2013)
Ishikawa, E., Mori, D. & Yamasaki, S. Recognition of mycobacterial lipids by immune receptors. Trends Immunol. 38, 66–76 (2017)
Bomar, L., Brugger, S. D., Yost, B. H., Davies, S. S. & Lemon, K. P. Corynebacterium accolens releases antipneumococcal free fatty acids from human nostril and skin surface triacylglycerols. MBio 7, e01725–e15 (2016)
Naik, S. et al. Commensal–dendritic-cell interaction specifies a unique protective skin immune signature. Nature 520, 104–108 (2015)
Linehan, J. L. et al. Non-classical immunity controls microbiota impact on skin immunity and tissue repair. Cell (in the press)
Clark, R. A. et al. The vast majority of CLA+ T cells are resident in normal skin. J. Immunol. 176, 4431–4439 (2006)
Metze, D. et al. Immunohistochemical demonstration of immunoglobulin A in human sebaceous and sweat glands. J. Invest. Dermatol. 92, 13–17 (1989)
Okada, T., Konishi, H., Ito, M., Nagura, H. & Asai, J. Identification of secretory immunoglobulin A in human sweat and sweat glands. J. Invest. Dermatol. 90, 648–651 (1988). This study used immunohistochemistry to show that secretory IgA was associated with human sweat glands, and was probably being actively transported in a way similar to the intestine. This study raises the question of how IgA on the skin influences microbiota composition and whether commensal microbes stimulate IgA secretion similarly to gut commensal flora.
Fagarasan, S. et al. Critical roles of activation-induced cytidine deaminase in the homeostasis of gut flora. Science 298, 1424–1427 (2002)
Macpherson, A. J., Hunziker, L., McCoy, K. & Lamarre, A. IgA responses in the intestinal mucosa against pathogenic and non-pathogenic microorganisms. Microbes Infect. 3, 1021–1035 (2001)
Kawamoto, S. et al. The inhibitory receptor PD-1 regulates IgA selection and bacterial composition in the gut. Science 336, 485–489 (2012)
van der Waaij, L. A., Limburg, P. C., Mesander, G. & van der Waaij, D. In vivo IgA coating of anaerobic bacteria in human faeces. Gut 38, 348–354 (1996)
Shroff, K. E., Meslin, K. & Cebra, J. J. Commensal enteric bacteria engender a self-limiting humoral mucosal immune response while permanently colonizing the gut. Infect. Immun. 63, 3904–3913 (1995)
Macpherson, A. J. & Uhr, T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science 303, 1662–1665 (2004)
Vossenkämper, A. et al. A role for gut-associated lymphoid tissue in shaping the human B cell repertoire. J. Exp. Med. 210, 1665–1674 (2013)
O’Riordan, K. & Lee, J. C. Staphylococcus aureus capsular polysaccharides. Clin. Microbiol. Rev. 17, 218–234 (2004)
Cheng, B. L. et al. Evaluation of serotypes 5 and 8 capsular polysaccharides in protection against Staphylococcus aureus in murine models of infection. Hum. Vaccin. Immunother. 13, 1609–1614 (2017)
Zimmermann, M. & Fischbach, M. A. A family of pyrazinone natural products from a conserved nonribosomal peptide synthetase in Staphylococcus aureus. Chem. Biol. 17, 925–930 (2010)
Wyatt, M. A. et al. Staphylococcus aureus nonribosomal peptide secondary metabolites regulate virulence. Science 329, 294–296 (2010)
Scharschmidt, T. C. et al. A wave of regulatory T cells into neonatal skin mediates tolerance to commensal microbes. Immunity 43, 1011–1021 (2015)
Scharschmidt, T. C. et al. Commensal microbes and hair follicle morphogenesis coordinately drive Treg migration into neonatal skin. Cell Host Microbe 21, 467–477.e5 (2017)
Lai, Y. et al. Commensal bacteria regulate Toll-like receptor 3-dependent inflammation after skin injury. Nat. Med. 15, 1377–1382 (2009)
Loesche, M. et al. Temporal stability in chronic wound microbiota is associated with poor healing. J. Invest. Dermatol. 137, 237–244 (2017)
Kalan, L. et al. Redefining the chronic-wound microbiome: fungal communities are prevalent, dynamic, and associated with delayed healing. MBio 7, e01058–e16 (2016)
Feingold, K. R. The outer frontier: the importance of lipid metabolism in the skin. J. Lipid Res. 50 (Suppl), S417–S422 (2009)
Brandner, J. M. Importance of tight junctions in relation to skin barrier function. Curr. Probl. Dermatol. 49, 27–37 (2016)
Natsuga, K. Epidermal barriers. Cold Spring Harb. Perspect. Med. 4, a018218 (2014)
McLean, W. H. I . Filaggrin failure—from ichthyosis vulgaris to atopic eczema and beyond. Br. J. Dermatol. 175 (Suppl 2), 4–7 (2016)
Madison, K. C. Barrier function of the skin: “la raison d’être” of the epidermis. J. Invest. Dermatol. 121, 231–241 (2003)
Cleaver, J. E. Common pathways for ultraviolet skin carcinogenesis in the repair and replication defective groups of xeroderma pigmentosum. J. Dermatol. Sci. 23, 1–11 (2000)
Martincorena, I. et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 348, 880–886 (2015)
Has, C. & Bruckner-Tuderman, L. The genetics of skin fragility. Annu. Rev. Genomics Hum. Genet. 15, 245–268 (2014)
Capell, B. C., Tlougan, B. E. & Orlow, S. J. From the rarest to the most common: insights from progeroid syndromes into skin cancer and aging. J. Invest. Dermatol. 129, 2340–2350 (2009)
Totté, J. E. et al. Prevalence and odds of Staphylococcus aureus carriage in atopic dermatitis: a systematic review and meta-analysis. Br. J. Dermatol. 175, 687–695 (2016)
Totté, J. E. E. et al. A systematic review and meta-analysis on Staphylococcus aureus carriage in psoriasis, acne and rosacea. Eur. J. Clin. Microbiol. Infect. Dis. 35, 1069–1077 (2016)
Huang, J. T., Abrams, M., Tlougan, B., Rademaker, A. & Paller, A. S. Treatment of Staphylococcus aureus colonization in atopic dermatitis decreases disease severity. Pediatrics 123, e808–e814 (2009)
Kobayashi, T . et al. Dysbiosis and Staphylococcus aureus colonization drives inflammation in atopic dermatitis. Immunity 42, 756–766 (2015). This study demonstrated potential mechanistic links between dysbiotic skin flora and inflammation in atopic dermatitis by using a mouse model of eczema with ADAM17 deficiency that recapitulates spontaneous development of dysbiotic flora and skin inflammation.
Leyden, J. J ., Marples, R. R. & Kligman, A. M. Staphylococcus aureus in the lesions of atopic dermatitis. Br. J. Dermatol. 90, 525–530 (1974). This was one of the first studies to demonstrate abundant Staphylococcus aureus colonization of patients with atopic dermatitis, even in areas of normal-appearing skin, and established the concept that colonizing microbes can have pathogenic effects without overt infection.
Conti, F. et al. Association between Staphylococcus aureus nasal carriage and disease phenotype in patients affected by systemic lupus erythematosus. Arthritis Res. Ther. 18, 177 (2016)
Nakagawa, S. et al. Staphylococcus aureus virulent PSMα peptides induce keratinocyte alarmin release to orchestrate IL-17-dependent skin inflammation. Cell Host Microbe 22, 667–677.e5 (2017)
Liu, H. et al. Staphylococcus aureus epicutaneous exposure drives skin inflammation via IL-36-mediated T cell responses. Cell Host Microbe 22, 653–666.e5 (2017)
Otto, M. Basis of virulence in community-associated methicillin-resistant Staphylococcus aureus. Annu. Rev. Microbiol. 64, 143–162 (2010)
de Haas, C. J. C. et al. Chemotaxis inhibitory protein of Staphylococcus aureus, a bacterial antiinflammatory agent. J. Exp. Med. 199, 687–695 (2004)
Luong, T. T. & Lee, C. Y. Overproduction of type 8 capsular polysaccharide augments Staphylococcus aureus virulence. Infect. Immun. 70, 3389–3395 (2002)
Uhlén, M. et al. Complete sequence of the staphylococcal gene encoding protein A. A gene evolved through multiple duplications. J. Biol. Chem. 259, 1695–1702 (1984)
Palmqvist, N., Patti, J. M., Tarkowski, A. & Josefsson, E. Expression of staphylococcal clumping factor A impedes macrophage phagocytosis. Microbes Infect. 6, 188–195 (2004)
Peschel, A. et al. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 274, 8405–8410 (1999)
Foster, T. J. Immune evasion by staphylococci. Nat. Rev. Microbiol. 3, 948–958 (2005)
Rooijakkers, S. H. M. et al. Immune evasion by a staphylococcal complement inhibitor that acts on C3 convertases. Nat. Immunol. 6, 920–927 (2005)
Sonesson, A. et al. Identification of bacterial biofilm and the Staphylococcus aureus derived protease, staphopain, on the skin surface of patients with atopic dermatitis. Sci. Rep. 7, 8689 (2017)
Zhang, L. J . et al. Innate immunity. Dermal adipocytes protect against invasive Staphylococcus aureus skin infection. Science 347, 67–71 (2015). This study showed that adipogenesis and adipocyte production of AMPs help to protect against Staphylococcus aureus infection via intradermal injection, demonstrating that in addition to keratinocytes and sebocytes, subcutaneous tissues can participate in the immue response to microbes.
Joshi, G. S., Spontak, J. S., Klapper, D. G. & Richardson, A. R. Arginine catabolic mobile element encoded speG abrogates the unique hypersensitivity of Staphylococcus aureus to exogenous polyamines. Mol. Microbiol. 82, 9–20 (2011)
Thurlow, L. R. et al. Functional modularity of the arginine catabolic mobile element contributes to the success of USA300 methicillin-resistant Staphylococcus aureus. Cell Host Microbe 13, 100–107 (2013)
Wentworth, A. B., Drage, L. A., Wengenack, N. L., Wilson, J. W. & Lohse, C. M. Increased incidence of cutaneous nontuberculous mycobacterial infection, 1980 to 2009: a population-based study. Mayo Clin. Proc. 88, 38–45 (2013)
Merritt, R. W. et al. Ecology and transmission of Buruli ulcer disease: a systematic review. PLoS Negl. Trop. Dis. 4, e911 (2010)
Houben, R. M. G. J. & Dodd, P. J. The global burden of latent tuberculosis infection: a re-estimation using mathematical modelling. PLoS Med. 13, e1002152 (2016)
World Health Organization. Global Tuberculosis Report 2016; http://apps.who.int/iris/bitstream/10665/250441/1/9789241565394-eng.pdf?ua=1 (2016)
Haley, C. A. Treatment of latent tuberculosis infection. Microbiol. Spectr. 5, TNMI7-0039–2016 (2017)
Sehgal, V. N. Leprosy. Dermatol. Clin. 12, 629–644 (1994)
Talhari, C., Talhari, S. & Penna, G. O. Clinical aspects of leprosy. Clin. Dermatol. 33, 26–37 (2015)
GBD 2016 Causes of Death Collaborators. Global, regional, and national age-sex specific mortality for 264 causes of death, 1980-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet 390, 1151–1210 (2017)
Wansbrough-Jones, M. & Phillips, R. Buruli ulcer: emerging from obscurity. Lancet 367, 1849–1858 (2006)
Marion, E . et al. Mycobacterial toxin induces analgesia in Buruli ulcer by targeting the angiotensin pathways. Cell 157, 1565–1576 (2014). This study shows that mycolactone, a virulence factor produced by the cutaneous pathogen Mycobacterium ulcerans , causes analgesia by directly binding to the angiotensin II receptor on nerve cells and triggering downstream potassium channel activation and resultant cell hyperpolarization.
Kashem, S. W. et al. Nociceptive sensory fibers drive interleukin-23 production from CD301b+ dermal dendritic cells and drive protective cutaneous immunity. Immunity 43, 515–526 (2015). This study shows that neurons can participate directly in the immune response to microbes. Cutaneous sensory neurons are directly activated by Candida albicans and subsequently stimulate dermal dendritic cells to produce IL-23, thus driving protective immunity by IL-17A-producing dermal T cells.
Chiu, I. M. et al. Bacteria activate sensory neurons that modulate pain and inflammation. Nature 501, 52–57 (2013)
Cardoso, V. et al. Neuronal regulation of type 2 innate lymphoid cells via neuromedin U. Nature 549, 277–281 (2017)
Bellono, N. W. et al. Enterochromaffin cells are gut chemosensors that couple to sensory neural pathways. Cell 170, 185–198 (2017)
Fung, T. C. et al. Lymphoid tissue-resident commensal bacteria promote members of the IL-10 cytokine family to establish mutualism. Immunity 44, 634–646 (2016)
Sonnenberg, G. F. et al. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science 336, 1321–1325 (2012)
Davis, J. M. & Ramakrishnan, L. The role of the granuloma in expansion and dissemination of early tuberculous infection. Cell 136, 37–49 (2009)
Horsburgh, C. R. J., Jr. Priorities for the treatment of latent tuberculosis infection in the United States. N. Engl. J. Med. 350, 2060–2067 (2004)
Adams, K. N. et al. Drug tolerance in replicating mycobacteria mediated by a macrophage-induced efflux mechanism. Cell 145, 39–53 (2011)
Schnappinger, D. et al. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198, 693–704 (2003)
Cunningham, A. F. & Spreadbury, C. L. Mycobacterial stationary phase induced by low oxygen tension: cell wall thickening and localization of the 16-kilodalton α-crystallin homolog. J. Bacteriol. 180, 801–808 (1998)
Rittershaus, E. S. C., Baek, S.-H. & Sassetti, C. M. The normalcy of dormancy: common themes in microbial quiescence. Cell Host Microbe 13, 643–651 (2013)
Bartek, I. L. et al. Mycobacterium tuberculosis Lsr2 is a global transcriptional regulator required for adaptation to changing oxygen levels and virulence. MBio 5, e01106–e01114 (2014)
Eoh, H. et al. Metabolic anticipation in Mycobacterium tuberculosis. Nat. Microbiol. 2, 201784 (2017)
Galagan, J. E. et al. The Mycobacterium tuberculosis regulatory network and hypoxia. Nature 499, 178–183 (2013)
Lin, P. L. et al. Sterilization of granulomas is common in active and latent tuberculosis despite within-host variability in bacterial killing. Nat. Med. 20, 75–79 (2014)
Hannigan, G. D. et al. The human skin double-stranded DNA virome: topographical and temporal diversity, genetic enrichment, and dynamic associations with the host microbiome. MBio 6, e01578–e15 (2015)
Hickman, H. D . et al. Anatomically restricted synergistic antiviral activities of innate and adaptive immune cells in the skin. Cell Host Microbe 13, 155–168 (2013). This study used intravital multiphoton microscopy to demonstrate that effector CD8+ T cells respond to cutaneous vaccinia virus infection by killing infected monocytes in the periphery but not infected keratinocytes in the center, thus highlighting the spatial complexity and specificity of immune cell dynamics in the skin.
Cush, S. S. et al. Locally produced IL-10 limits cutaneous vaccinia virus spread. PLoS Pathog. 12, e1005493 (2016)
Carbone, F. R. Tissue-resident memory T cells and fixed immune surveillance in nonlymphoid organs. J. Immunol. 195, 17–22 (2015)
Mueller, S. N., Gebhardt, T., Carbone, F. R. & Heath, W. R. Memory T cell subsets, migration patterns, and tissue residence. Annu. Rev. Immunol. 31, 137–161 (2013)
Lanas, A. & Chan, F. K. L. Peptic ulcer disease. Lancet 390, 613–624 (2017)
Kalisperati, P. et al. Inflammation, DNA damage, Helicobacter pylori and gastric tumorigenesis. Front. Genet. 8, 20 (2017)
Peek, R. M., Jr & Blaser, M. J. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat. Rev. Cancer 2, 28–37 (2002)
Kienesberger, S. et al. Gastric Helicobacter pylori infection affects local and distant microbial populations and host responses. Cell Reports 14, 1395–1407 (2016)
Lehman, H. Skin manifestations of primary immune deficiency. Clin. Rev. Allergy Immunol. 46, 112–119 (2014)
Barnard, E., Shi, B., Kang, D., Craft, N. & Li, H. The balance of metagenomic elements shapes the skin microbiome in acne and health. Sci. Rep. 6, 39491 (2016)
Agak, G. W. et al. Propionibacterium acnes Induces an IL-17 response in acne vulgaris that is regulated by vitamin A and vitamin D. J. Invest. Dermatol. 134, 366–373 (2014)
Fitz-Gibbon, S. et al. Propionibacterium acnes strain populations in the human skin microbiome associated with acne. J. Invest. Dermatol. 133, 2152–2160 (2013)
Ring, H. C. et al. The follicular skin microbiome in patients with hidradenitis suppurativa and healthy controls. JAMA Dermatol. 153, 897–905 (2017)
Wollenberg, M. S. et al. Propionibacterium-produced coproporphyrin III induces Staphylococcus aureus aggregation and biofilm formation. MBio 5, e01286–e14 (2014)
Shu, M. et al. Fermentation of Propionibacterium acnes, a commensal bacterium in the human skin microbiome, as skin probiotics against methicillin-resistant Staphylococcus aureus. PLoS One 8, e55380 (2013)
Barton, E. S. et al. Herpesvirus latency confers symbiotic protection from bacterial infection. Nature 447, 326–329 (2007)
Yager, E. J. et al.. γ-Herpesvirus-induced protection against bacterial infection is transient. Viral Immunol. 22, 67–72 (2009)
Perry, S. et al. Infection with Helicobacter pylori is associated with protection against tuberculosis. PLoS One 5, e8804 (2010)
Arnold, I. C. et al. Helicobacter pylori infection prevents allergic asthma in mouse models through the induction of regulatory T cells. J. Clin. Invest. 121, 3088–3093 (2011)
Cheung, G. Y. C. & Otto, M. Understanding the significance of Staphylococcus epidermidis bacteremia in babies and children. Curr. Opin. Infect. Dis. 23, 208–216 (2010)
Rohlke, F. & Stollman, N. Fecal microbiota transplantation in relapsing Clostridium difficile infection. Therap. Adv. Gastroenterol. 5, 403–420 (2012)
Panigrahi, P . et al. A randomized synbiotic trial to prevent sepsis among infants in rural India. Nature 548, 407–412 (2017). This study was the first, to our knowledge, to use an oral synbiotic ( Lactobacillus plantarum and fructooligosaccharide) to promote effective gut colonization of the inoculated bacterium and reduce neonatal sepsis.
Nakatsuji, T. et al. Sebum free fatty acids enhance the innate immune defense of human sebocytes by upregulating β-defensin-2 expression. J. Invest. Dermatol. 130, 985–994 (2010)
Falchook, G. S. et al. Responses of metastatic basal cell and cutaneous squamous cell carcinomas to anti-PD1 monoclonal antibody REGN2810. J. Immunother. Cancer 4, 70 (2016)
Morris, V. K. et al. Nivolumab for previously treated unresectable metastatic anal cancer (NCI9673): a multicentre, single-arm, phase 2 study. Lancet Oncol. 18, 446–453 (2017)
Mahoney, K. M., Freeman, G. J. & McDermott, D. F. The next immune-checkpoint inhibitors: PD-1/PD-L1 blockade in melanoma. Clin. Ther. 37, 764–782 (2015)
Cassler, N. M. & Brownell, I. PD-1 checkpoint blockade is an emerging treatment for Merkel cell carcinoma. Br. J. Dermatol. 176, 18 (2017)
Sivan, A . et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 350, 1084–1089 (2015). This study and the next one demonstrated that the gut microbial composition can alter immunotherapies at distant sites, suggesting that microbe–immune interactions at barrier sites can have far-reaching effects at other barrier sites or systemically.
Vétizou, M. et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 350, 1079–1084 (2015)
Golden, J. B. et al. Chronic, not acute, skin-specific inflammation promotes thrombosis in psoriasis murine models. J. Transl. Med. 13, 382 (2015)
Santilli, S. et al. Visualization of atherosclerosis as detected by coronary artery calcium and carotid intima-media thickness reveals significant atherosclerosis in a cross-sectional study of psoriasis patients in a tertiary care center. J. Transl. Med. 14, 217 (2016)
Wang, Y . et al. Chronic skin-specific inflammation promotes vascular inflammation and thrombosis. J. Invest. Dermatol. 132, 2067–2075 (2012). This study used a mouse model of psoriasis (keratinocyte-specific Tie2 transgene expression) to mechanistically link skin inflammation to the development of resultant aortic root inflammation and also to show that subsequent treatment of the skin disease can eliminate not only skin inflammation but also systemic vascular inflammation.
Evensen, K. et al. Increased subclinical atherosclerosis in patients with chronic plaque psoriasis. Atherosclerosis 237, 499–503 (2014)
Tomura, M. et al. Activated regulatory T cells are the major T cell type emigrating from the skin during a cutaneous immune response in mice. J. Clin. Invest. 120, 883–893 (2010)
Tomura, M. et al. Tracking and quantification of dendritic cell migration and antigen trafficking between the skin and lymph nodes. Sci. Rep. 4, 6030 (2014)
Geherin, S. A. et al. The skin, a novel niche for recirculating B cells. J. Immunol. 188, 6027–6035 (2012). This study was one of the first to demonstrate that B cells actively traffic in and out of the skin, even in uninflamed skin, suggesting that B cells play an active and previously underappreciated role in skin homeostasis.
Belkaid, Y. & Segre, J. A. Dialogue between skin microbiota and immunity. Science 346, 954–959 (2014)
Gallo, R. L. & Hooper, L. V. Epithelial antimicrobial defence of the skin and intestine. Nat. Rev. Immunol. 12, 503–516 (2012)
Nithya, S., Radhika, T. & Jeddy, N. Loricrin—an overview. J. Oral Maxillofac. Pathol. 19, 64–68 (2015)
Iguchi, A. et al. A complete view of the genetic diversity of the Escherichia coli O-antigen biosynthesis gene cluster. DNA Res. 22, 101–107 (2015)
Acknowledgements
We apologize for not having cited all papers relevant to this expanding field of research (in particular, older literature) because of space constraints and editorial limits. This work was supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases (Y.B.), DP1 DK113598 (M.A.F.), R01 DK110174 (M.A.F.), an HHMI-Simons Faculty Scholars Award (M.A.F.), a Fellowship for Science and Engineering from the David and Lucile Packard Foundation (M.A.F.), a Burroughs Wellcome Investigators in the Pathogenesis of Infectious Disease Award (M.A.F.) and the Dermatology Foundation (Y.E.C.).
Author information
Authors and Affiliations
Contributions
Y.E.C., M.A.F. and Y.B. conceptualized the article structure, content, and figures, and wrote and edited the manuscript and figures.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks T. Scharschmidt and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Chen, Y., Fischbach, M. & Belkaid, Y. Skin microbiota–host interactions. Nature 553, 427–436 (2018). https://doi.org/10.1038/nature25177
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature25177
This article is cited by
-
Surface nanocoating of bacteria as a versatile platform to develop living therapeutics
Nature Protocols (2024)
-
High-throughput identification and quantification of bacterial cells in the microbiota based on 16S rRNA sequencing with single-base accuracy using BarBIQ
Nature Protocols (2024)
-
Dermal injury drives a skin to gut axis that disrupts the intestinal microbiome and intestinal immune homeostasis in mice
Nature Communications (2024)
-
Metabolomic profiling of wild rooibos (Aspalathus linearis) ecotypes and their antioxidant-derived phytopharmaceutical potential
Metabolomics (2024)
-
Unveiling the mechanism of essential oil action against skin pathogens: from ancient wisdom to modern science
Archives of Microbiology (2024)