Abstract
We have sequenced the genomes of 110 small cell lung cancers (SCLC), one of the deadliest human cancers. In nearly all the tumours analysed we found bi-allelic inactivation of TP53 and RB1, sometimes by complex genomic rearrangements. Two tumours with wild-type RB1 had evidence of chromothripsis leading to overexpression of cyclin D1 (encoded by the CCND1 gene), revealing an alternative mechanism of Rb1 deregulation. Thus, loss of the tumour suppressors TP53 and RB1 is obligatory in SCLC. We discovered somatic genomic rearrangements of TP73 that create an oncogenic version of this gene, TP73Δex2/3. In rare cases, SCLC tumours exhibited kinase gene mutations, providing a possible therapeutic opportunity for individual patients. Finally, we observed inactivating mutations in NOTCH family genes in 25% of human SCLC. Accordingly, activation of Notch signalling in a pre-clinical SCLC mouse model strikingly reduced the number of tumours and extended the survival of the mutant mice. Furthermore, neuroendocrine gene expression was abrogated by Notch activity in SCLC cells. This first comprehensive study of somatic genome alterations in SCLC uncovers several key biological processes and identifies candidate therapeutic targets in this highly lethal form of cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
Primary accessions
Gene Expression Omnibus
Data deposits
Affymetrix SNP 6.0, whole-genome, and transcriptome sequencing on human specimen have been deposited at the European Genome-phenome Archive under the accession code EGAS00001000925. Whole-exome and whole-genome sequencing data of murine SCLC tumors can be accessed through (http://www.translational-genomics.uni-koeln.de/scientific-resources/). Microarray data on mouse cell lines is accessible through Gene Expression Omnibus (GEO) accession number GSE69091.
References
van Meerbeeck, J. P., Fennell, D. A. & De Ruysscher, D. K. Small-cell lung cancer. Lancet 378, 1741–1755 (2011)
Peifer, M. et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nature Genet. 44, 1104–1110 (2012)
Rudin, C. M. et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nature Genet. 44, 1111–1116 (2012)
Takahashi, T. et al. p53: a frequent target for genetic abnormalities in lung cancer. Science 246, 491–494 (1989)
Wistuba, I. I., Gazdar, A. F. & Minna, J. D. Molecular genetics of small cell lung carcinoma. Semin. Oncol. 28, 3–13 (2001)
Horowitz, J. M. et al. Frequent inactivation of the retinoblastoma anti-oncogene is restricted to a subset of human tumor cells. Proc. Natl Acad. Sci. USA 87, 2775–2779 (1990)
Mori, N. et al. Variable mutations of the RB gene in small-cell lung carcinoma. Oncogene 5, 1713–1717 (1990)
Meuwissen, R. et al. Induction of small cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model. Cancer Cell 4, 181–189 (2003)
Schaffer, B. E. et al. Loss of p130 accelerates tumor development in a mouse model for human small-cell lung carcinoma. Cancer Res. 70, 3877–3883 (2010)
McFadden, D. G. et al. Genetic and clonal dissection of murine small cell lung carcinoma progression by genome sequencing. Cell 156, 1298–1311 (2014)
Imielinski, M. et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 150, 1107–1120 (2012)
Zhang, J. et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science 346, 256–259 (2014)
de Bruin, E. C. et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 346, 251–256 (2014)
Futreal, P. A. et al. A census of human cancer genes. Nature Rev. Cancer 4, 177–183 (2004)
Forbes, S. A. et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 43, D805–D811 (2015)
Lawrence, M. S. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013)
Yokomizo, A. et al. PTEN/MMAC1 mutations identified in small cell, but not in non-small cell lung cancers. Oncogene 17, 475–479 (1998)
Holderfield, M., Deuker, M. M., McCormick, F. & McMahon, M. Targeting RAF kinases for cancer therapy: BRAF-mutated melanoma and beyond. Nature Rev. Cancer 14, 455–467 (2014)
Hibi, K. et al. Coexpression of the stem cell factor and the c-kit genes in small-cell lung cancer. Oncogene 6, 2291–2296 (1991)
Hirota, S. et al. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science 279, 577–580 (1998)
Shibata, T., Kokubu, A., Tsuta, K. & Hirohashi, S. Oncogenic mutation of PIK3CA in small cell lung carcinoma: a potential therapeutic target pathway for chemotherapy-resistant lung cancer. Cancer Lett. 283, 203–211 (2009)
Angeloni, D. et al. Analysis of a new homozygous deletion in the tumor suppressor region at 3p12.3 reveals two novel intronic noncoding RNA genes. Genes Chromosom. Cancer 45, 676–691 (2006)
Kovatich, A. et al. Molecular alterations to human chromosome 3p loci in neuroendocrine lung tumors. Cancer 83, 1109–1117 (1998)
Weiss, J. et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci. Transl. Med. 2, 62ra93 (2010)
Korbel, J. O. & Campbell, P. J. Criteria for inference of chromothripsis in cancer genomes. Cell 152, 1226–1236 (2013)
Knudson, A. G. Two genetic hits (more or less) to cancer. 1, Nature Rev. Genet. 1, 157–162 (2001)
Berger, A. H., Knudson, A. G. & Pandolfi, P. P. A continuum model for tumour suppression. Nature 476, 163–169 (2011)
Beasley, M. B. et al. The P16/cyclin D1/Rb pathway in neuroendocrine tumors of the lung. Hum. Pathol. 34, 136–142 (2003)
Northcott, P. A. et al. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature 511, 428–434 (2014)
The Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 (2012)
Pitkänen, E., Cajuso, T., Katainen, R., Kaasinen, E. & Välimäki, N. Frequent L1 retrotranspositions originating from TTC28 in colorectal cancer. Oncotarget 5, 853–859 (2014)
Helman, E., Lawrence, M. L. & Stewart, C. Somatic retrotransposition in human cancer revealed by whole-genome and exome sequencing. Genome Res. 24, 1053–1063 (2014)
Jancalek, R. The role of the TP73 gene and its transcripts in neuro-oncology. Br. J. Neurosurg. 28, 598–605 (2014)
Tannapfel, A. et al. Autonomous growth and hepatocarcinogenesis in transgenic mice expressing the p53 family inhibitor DNp73. Carcinogenesis 29, 211–218 (2008)
Venkatanarayan, A. et al. IAPP-driven metabolic reprogramming induces regression of p53-deficient tumours in vivo. Nature 517, 626–630 (2015)
Falix, F. A., Aronson, D. C., Lamers, W. H. & Gaemers, I. C. Possible roles of DLK1 in the Notch pathway during development and disease. Biochim. Biophys. Acta 1822, 988–995 (2012)
Ball, D. W. Achaete-scute homolog-1 and Notch in lung neuroendocrine development and cancer. Cancer Lett. 204, 159–169 (2004)
Augustyn, A. et al. ASCL1 is a lineage oncogene providing therapeutic targets for high-grade neuroendocrine lung cancers. Proc. Natl Acad. Sci. USA 111, 14788–14793 (2014)
Sriuranpong, V. et al. Notch signaling induces cell cycle arrest in small cell lung cancer cells Cancer Res. 1, 3200–3205 (2001)
Wael, H. et al. Notch1 signaling controls cell proliferation, apoptosis and differentiation in lung carcinoma. Lung Cancer 85, 131–140 (2014)
Rangarajan, A. et al. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 20, 3427–3436 (2001)
Qi, R. et al. Notch1 signaling inhibits growth of human hepatocellular carcinoma through induction of cell cycle arrest and apoptosis. Cancer Res. 63, 8323–8329 (2003)
Iwakawa, R. et al. Genome-wide identification of genes with amplification and/or fusion in small cell lung cancer. Genes Chromosomes Cancer 52, 802–816 (2013)
Seidel, D. A genomics-based classification of human lung tumors. Sci. Transl. Med. 5, 209ra153 (2013)
Lafkas, D. et al. Notch3 marks clonogenic mammary luminal progenitor cells in vivo. J. Cell Biol. 203, 47–56 (2013)
Muzumdar, M. D., Tasic, B., Miyamichi, K., Li, L. & Luo, L. A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 (2007)
Lu, X., Thomas, R. K. & Peifer, M. CGARS: cancer genome analysis by rank sums. Bioinformatics 30, 1295–1296 (2014)
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009)
Fernandez-Cuesta, L. et al. Frequent mutations in chromatin-remodelling genes in pulmonary carcinoids. Nat. Commun. 5, 3518 (2014)
Jahchan, N. S. et al. A drug repositioning approach identifies tricyclic antidepressants as inhibitors of small cell lung cancer and other neuroendocrine tumors. Cancer Discov. 3, 1364–1377 (2013)
Acknowledgements
We are grateful to all the patients who contributed their tumour specimens. We thank the computing center of the University of Cologne (RRZK) for providing the CPU time on the DFG-funded supercomputer ‘CHEOPS’, as well as for the support. We thank S. Artavanis-Tsakonas and S. Fre for the gift of the mice with inducible NICD expression. We thank C. Nguyen, J. Berg, J. Heuckmann, F. Malchers, C. Lovely and A. Bernschein for scientific discussions and advice. We thank Genentech/gRED for providing raw sequencing data from a previously published study3. Some tumors in these studies were provided by the LungBiobank Heidelberg, member of the NCT-Tissue bank, the biomaterial bank Heidelberg and the biobank platform of the German Center for Lung Research, Heidelberg, Germany. This work was supported by the German Cancer Aid (Deutsche Krebshilfe) as part of the small cell lung cancer genome sequencing consortium (grant ID: 109679 to R.K.T., M.P., R.B., P.N., M.V. and S.A.H.). Further support was provided by the Korea Research Foundation (KRF 2011-0030105; grant to S.J.J.). Additional funding was provided by the NIH (5R01CA114102-08 to J.S.), the German Ministry of Science and Education (BMBF) as part of the NGFNplus program (grant 01GS08101 to R.K.T., J.W. and P.N.) and as part of the e:Med program (grant no. 01ZX1303A to R.K.T., J.W., C.R., R.B. and M.P. and grant no. 01ZX1406 to M.P.), by the Deutsche Forschungsgemeinschaft (DFG; through TH1386/3-1 to R.K.T and KFO-286 to P.N.), by the German federal state North Rhine Westphalia (NRW), by the European Union (European Regional Development Fund: Investing In Your Future) as part of the PerMed NRW initiative (grant 005-1111-0025 to R.K.T., J.W. and R.B.), by SFB832 (TP6 to R.K.T., TP5 to L.C.H.), by the Deutsche Krebshilfe as part of the Oncology Centers of Excellence funding program (to R.B., R.K.T. and M.S.), by the EU-Framework program CURELUNG (HEALTH-F2-2010-258677 to R.K.T., J.W., J.K.F., L.R., M.S.C. and E.B.), by Stand Up To Cancer — American Association of Cancer Research Innovative Research Grant (SU2C-AACR-IR60109 to R.K.T.), by the German Consortium for Translational Cancer Research (DKTK) Joint Funding program, by the National Cancer Center Research and Development Fund (NCC Biobank: 23A-1, to T.K., J.Y. and R.I.), by the Italian Ministry of Health (Ricerca Corrente RC1303LO57 and GR program 2010-2316264 to L.A.M.), by the Roy Castle Lung Cancer Foundation UK (to J.K.F.), by the AIRC/MGAF grant 12983 (to L.A.M.) and by A*STAR in Singapore (scholarship to J.S.L.). J.S. is the Harriet and Mary Zelencik Scientist in Children’s Cancer and Blood Diseases.
Author information
Authors and Affiliations
Contributions
J.G., J.S.L., M.P., J.S. and R.K.T. conceived the project, analysed and interpreted the data, and wrote the manuscript. J.G., J.S.L., J.S., F.L., R.M., S.P., D.E., B.Pü, M.S.W., J.O.K., J.A., C.B., M.B. and P.S. designed experiments. J.G., J.S.L., I.D., C.M., A.T., R.M., S.-M.C., D.K., D.E., I.V., D.S., B.Pi, P.S., C.B., P.M.S. and M.Bog performed experiments. J.G., M.P., J.S.L., S.A.H., M.V., M.S.W., J.O.K., Y.C., X.L., D.V., M.W., N.H. and M.Bos performed data analysis. E.B., W.D.T., R.B., L.H., L.O., S.P., S.J.J. and G.K. performed pathology review. E.B., W.D.T., L.O., A.N.K., Y.Y. and V.T. conducted further immunohistochemistry studies. F.L., L.F.-C., G.B., S.M., D.S., V.A., U.L., T.Z., S.A., M.H., J.W., P.N. and C.R. helped with logistics. S.J.J., N.S.J., K.-S.P., D.Y., J.Y.,T.K., R.I., K.T., M.N., T.M., H.H., P.A.S., I.P., Y.C., A.S., C.-M.C., Y.-H.K., P.P.M., Y.Z., D.J., M.K., G.M.W., P.A.R., B.S., I.K., M.L., L.A.M, A.l.T., J.K.F., M.J., J.Kn., E.C.-V., L.R., U.P., O.-T.B., M.L.-I., E.T., J.Kö., M.Sc, J.B., M.Sa, M.S.-C., H.B.S., Y.Y., S.P., L.H., R.B. and E.B. contributed with murine and human tissue samples.
Corresponding authors
Ethics declarations
Competing interests
Competing financial interests. R.K.T. is a founder and shareholder of NEO New Oncology AG, received commercial research grants from AstraZeneca, EOS and Merck KgaA and honoraria from AstraZeneca, Bayer, NEO New Oncology AG, Boehringer Ingelheim, Clovis Oncology, Daiichi-Sankyo, Eli Lilly, Johnson & Johnson, Merck KgaA, MSD, Puma, Roche and Sanofi. P.N. is a founder, CEO and shareholder of ATLAS Biolabs. ATLAS Biolabs is a service provider for genomic analyses. M.P. is a founder and shareholder of NEO New Oncology AG and receives consultation fees from NEO New Oncology AG. J.G. has been consulting for NEO New Oncology AG and G1 Therapeutics.
Extended data figures and tables
Extended Data Figure 1 Genomic analyses in SCLC tumours.
a, Schematic detailing the genomic study and number of samples as well as various steps of analyses for the identification of candidate genes in SCLC. b, Illustration of the number of samples analysed in this study.
Extended Data Figure 2 Clinical molecular-correlation analyses.
a, Survival analysis of SCLC patients based on clinical stage and treatment options (surgery and/or chemotherapy). Statistical significance was determined by log-rank test. b, Analyses of clinical stage and smoking status and the respective effect on number and type of mutations, as well as mutational subclonality in tumours. Statistical significance was determined by Kruskal–Wallis analysis.
Extended Data Figure 3 Genomic characterization of SCLC tumours.
a, Purity and ploidy determined in SCLC tumours by whole-genome sequencing presented as dot density plots showing median and the interquartile range (IQR) b, Subclonal architecture of SCLC in comparison to lung adenocarcinoma (AD). Whole-genome sequencing data of SCLC and of adenocarcinoma (n = 15)11 was analysed for the presence of subclonal populations using clustering of the derived cancer cell fraction (CCF) of all single nucleotide mutations. To compare the emerging subclonal structure, we derived a subclonality score that takes into account the CCF of each sub-population as well as its mutational burden (see Methods). In order to prevent the low sequencing coverage (35× for SCLC and 63× for AD) from causing a systematic underrepresentation of the subclonal diversity in the mutation calls, we computed the contribution of a single read to the CCF on genome-wide average. After systematically determining a threshold within the average increase of CCF per read values (see Methods for details), we determined the group of samples for which a reliable estimation of the subclonality score is not possible (grey area). The subclonality scores of the remaining SCLC cases were then compared to those of the adenocarcinoma cases (P = 0.000232; Mann–Whitney test). c, Schematic representation of candidate genes with significant clustering of mutations in respective protein domains. Somatic mutations and genomic translocations are mapped to the respective protein regions. Hotspot mutations are highlighted in red. d, e, Genomic alterations in the RB1 family proteins p107 (RBL1) and p130 (RBL2) (d), and in KIT and PIK3CA (e). Somatic mutations in therapeutic target genes are listed and mapped to the protein domains of KIT and PIK3CA. Mutations with potential therapeutic implications are highlighted in red.
Extended Data Figure 4 Clinical molecular-correlations of significantly mutated genes.
a, Survival analysis of SCLC patients based on the status of CREBBP/EP300, TP73 or NOTCH alterations. Statistical significance was determined by log-rank test. b, Analysis of CREBBP/EP300, TP73 and NOTCH alterations and their effect on clinical and genetic parameters. Statistical significance was analysed by multinomial logistic regression.
Extended Data Figure 5 Significant somatic copy number alterations in SCLC.
a, Deletions of the chromosomal arm 3p point to the 3p14 (FHIT) and 3p12 (ROBO1) locus. b, Expression analyses of genes encoded on the 3p14.3–3p14.2 and 3p12.2–3p12.2 locus. Histogram displaying the expression of samples with focal deletions (blue) and samples without any copy number alterations (white). Mean and standard error of the mean is plotted for each gene in each group. Significant differences were determined by Mann–Whitney test; *P < 0.05; **P < 0.01. c, d, Focal deletions of the CDKN2A (c) and focal amplifications of IRS2 (d) were found on chromosome 9 and 13, respectively. The copy number (CN) states were computed from SNP array (SNP 6.0) and from whole-genome sequencing (WGS) data. The samples are sorted according to their amplitude of deletions or amplifications. e, Amplifications of IRS2 were determined by FISH analysis. IRS amplifications were quantified based on the ratio of red signals (IRS2-specific probe) to green signals (centromere probe for chromosome 13). Lymphocyte spreads and SCLC tumours without detectable IRS2 amplifications served as negative controls. Scale bar, 100 µm.
Extended Data Figure 6 TP53 and RB1 alterations in SCLC.
a, Distribution of somatic mutations in TP53 and RB1 according to the colour panel provided. b, c, Complex genomic rearrangements in RB1 showing homozygous deletions of exon 1 (b) or inversions within the RB1 gene (c). d, e, Annotated silent or missense mutations in RB1 occur at intron-exon junctions resulting in alternative splicing, intron retention (d) or exon skipping events (e). The coverage at the respective exon junctions is quantified as RPKM values. Sample S02194 is not holding any mutations at intron–exon junctions and is displayed as an example for unaltered splicing of RB1.
Extended Data Figure 7 Chromothripsis in human SCLC.
a, Circos plot of the chromothripsis sample S02353 showing intra- and interchromosomal rearrangements between chromosome 3 and 11. The integral copy number state (iCN) is plotted as a heatmap and assigned to the respective chromosomal regions. The chromosomal context of CCND1 (on chromosome 11) is highlighted. b, Circos plots displaying fusion transcripts identified in the SCLC chromothripsis cases (Supplementary Table 12) are represented as blue (S02297) or red (S02353) lines for genes located on chromosome 3 and 11. c, Immunohistochemistry staining for p53, p14 (ARF) and p16 on FFPE material of the chromothripsis sample S02297. Original magnification, ×400.
Extended Data Figure 8 Recurrent genomic translocations in SCLC.
a, Recurrent genomic translocations (n = 14) affecting chromosome 22 are illustrated as a Circos plot highlighting the respective rearrangements as red connecting lines. b, Breakpoints in chromosome 22 map to intron 1 of TTC28 and cluster downstream of the LINE1 (L1Hs) retrotransposon. Each arrow indicates the sample and the respective chromosomal position the segment translocates to. c, Schematic representation of the TP73 locus (hg19) describing complex intrachromosomal rearrangements of TP73 identified for S02397 and S02243. Recurrent somatic mutations identified in Fig. 1a are mapped to the respective exons. d, Validation of somatic TP73 translocations. Genomic regions involved in the TP73 rearrangements were amplified in matched normal (N) and tumour (T) samples. The expected band size is indicated in brackets. The respective PCR products were subjected to Sanger sequencing to confirm the genomic breakpoint. e, Copy-number state of the TP73 gene in samples involved in genomic translocations.
Extended Data Figure 9 Transcriptome profile of human SCLC tumours.
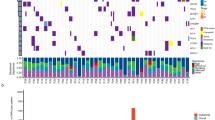
a, Unsupervised hierarchical clustering of transcriptome sequencing data of 69 SCLC specimens as described in Fig. 4a. Each sample is annotated for the genomic alterations described in Fig. 1. Black filled boxes describe the presence of a genomic event. b, Expression values of CHGA, GRP, ASCL1 and DLK1 (FPKM) are represented as dot density plots for the subgroups identified in (a). Red lines highlight the median value for each group. c, Expression values of the neuroendocrine markers SYP (synaptophysin) and NCAM1 (CD56) plotted as scatter plots for all SCLC samples. Green lines indicate thresholds for no expression (FPKM<1).
Extended Data Figure 10 Notch is a tumour suppressor in SCLC regulating neuroendocrine differentiation.
a, Somatic mutations identified in NOTCH3 and NOTCH4 are mapped to the protein domains. Damaging mutations are highlighted in red. Mutations found in murine SCLC tumours are highlighted in blue. b, Quantification of tumour lesions and per cent tumour area to lung in TKO (n = 5) and TKO;N1ICD (n = 4) mice 3 months after Ad-Cre instillation. Statistical significance was determined by two-tailed unpaired Student’s t-test. c, Representative immunohistochemistry for GFP or tdTomato in lungs from TKO;Rosa26mT/mG mice approximately 6 months after tumour induction. Left scale bar, 500 μm; right and middle: scale bar, 50 μm. d, Representative immunostaining for Notch2 in lungs from TKO;Rosa26N2ICD mice approximately 6 months after tumour induction. Left scale bar, 500 μm; right scale bar, 50 μm. e, Quantification of the per cent recombination at the Rosa26 locus in TKO;Rosa26mT/mG (n = 6) and TKO;Rosa26N2ICD mice (n = 10; two-tailed unpaired Student’s t-test). f, Cell viability assay of the human SCLC cell line NJH29 transfected with a N1ICD (Notch1) expression plasmid or empty vector control (Ctrl) (3 independent biological replicas with 3 technical replicas each). Fold growth was normalized to day 0; representative images were taken on day 6. Scale bar, 50 μm. g, Immunohistochemistry staining in FFPE embedded tissues of TKO and TKO;N2ICD mice. Scale bar, 50 μm. h, Quantitative RT–PCR validation of Notch1 induction and the expression of common Notch target genes after N1ICD transfection in murine SCLC cells (three biological replicas; two-tailed paired Student’s t-test). i, Mouse SCLC cells transfected with control or N1ICD (Notch1) were analysed 48 h later by gene expression microarrays. Gene Set Enrichment Analysis (GSEA) was performed on these data; selected significant gene sets are displayed. j, k, EdU analysis of mouse (j) and human (k) SCLC cells (three independent biological replicas with three technical replicas each; two-tailed paired Student’s t-test). *P < 0.05; **P < 0.01; ***P < 0.001. Data are represented as mean ± s.d.
Supplementary information
Supplementary Data
This file contains Supplementary Tables 1-15. (XLSX 32236 kb)
Rights and permissions
About this article
Cite this article
George, J., Lim, J., Jang, S. et al. Comprehensive genomic profiles of small cell lung cancer. Nature 524, 47–53 (2015). https://doi.org/10.1038/nature14664
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14664