Abstract
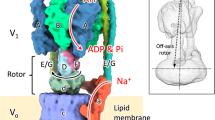
Eukaryotic vacuolar H+-ATPases (V-ATPases) are rotary enzymes that use energy from hydrolysis of ATP to ADP to pump protons across membranes and control the pH of many intracellular compartments. ATP hydrolysis in the soluble catalytic region of the enzyme is coupled to proton translocation through the membrane-bound region by rotation of a central rotor subcomplex, with peripheral stalks preventing the entire membrane-bound region from turning with the rotor. The eukaryotic V-ATPase is the most complex rotary ATPase: it has three peripheral stalks, a hetero-oligomeric proton-conducting proteolipid ring, several subunits not found in other rotary ATPases, and is regulated by reversible dissociation of its catalytic and proton-conducting regions1,2. Studies of ATP synthases, V-ATPases, and bacterial/archaeal V/A-ATPases have suggested that flexibility is necessary for the catalytic mechanism of rotary ATPases3,4,5, but the structures of different rotational states have never been observed experimentally. Here we use electron cryomicroscopy to obtain structures for three rotational states of the V-ATPase from the yeast Saccharomyces cerevisiae. The resulting series of structures shows ten proteolipid subunits in the c-ring, setting the ATP:H+ ratio for proton pumping by the V-ATPase at 3:10, and reveals long and highly tilted transmembrane α-helices in the a-subunit that interact with the c-ring. The three different maps reveal the conformational changes that occur to couple rotation in the symmetry-mismatched soluble catalytic region to the membrane-bound proton-translocating region. Almost all of the subunits of the enzyme undergo conformational changes during the transitions between these three rotational states. The structures of these states provide direct evidence that deformation during rotation enables the smooth transmission of power through rotary ATPases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
Electron Microscopy Data Bank
Protein Data Bank
Data deposits
Cryo-EM maps have been deposited in the Electron Microscopy Data Bank under accession numbers EMD-6284, EMD-6285, and EMD-6286. Atomic models have been deposited in the Protein Data Bank under accession numbers 3J9T, 3J9U, and 3J9V.
References
Sumner, J. P. et al. Regulation of plasma membrane V-ATPase activity by dissociation of peripheral subunits. J. Biol. Chem. 270, 5649–5653 (1995)
Kane, P. M. Disassembly and reassembly of the yeast vacuolar H+-ATPase in vivo. J. Biol. Chem. 270, 17025–17032 (1995)
Pänke, O., Cherepanov, D. A., Gumbiowski, K., Engelbrecht, S. & Junge, W. Viscoelastic dynamics of actin filaments coupled to rotary F-ATPase: angular torque profile of the enzyme. Biophys. J. 81, 1220–1233 (2001)
Stewart, A. G., Lee, L. K., Donohoe, M., Chaston, J. J. & Stock, D. The dynamic stator stalk of rotary ATPases. Nature Commun. 3, 687 (2012)
Zhou, M. et al. Ion mobility–mass spectrometry of a rotary ATPase reveals ATP-induced reduction in conformational flexibility. Nature Chem. 6, 208–215 (2014)
Walker, J. E. ATP synthesis by rotary catalysis (Nobel Lecture). Angew. Chem. Int. Edn 37, 2309–2319 (1998)
Walker, J. E. Keilin Memorial Lecture. The ATP synthase: the understood, the uncertain and the unknown. Biochem. Soc. Trans. 41, 1–16 (2013)
Arai, S. et al. Rotation mechanism of Enterococcus hirae V1-ATPase based on asymmetric crystal structures. Nature 493, 703–707 (2013)
Benlekbir, S., Bueler, S. A. & Rubinstein, J. L. Structure of the vacuolar-type ATPase from Saccharomyces cerevisiae at 11-Å resolution. Nature Struct. Mol. Biol. 19, 1356–1362 (2012)
Rawson, S. et al. Structure of the vacuolar H+-ATPase rotary motor reveals new mechanistic insights. Structure 23, 461–471 (2015)
Amunts, A. et al. Structure of the yeast mitochondrial large ribosomal subunit. Science 343, 1485–1489 (2014)
Trabuco, L. G., Villa, E., Mitra, K., Frank, J. & Schulten, K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure 16, 673–683 (2008)
Hirata, R., Graham, L. A., Takatsuki, A., Stevens, T. H. & Anraku, Y. VMA11 and VMA16 encode second and third proteolipid subunits of the Saccharomyces cerevisiae vacuolar membrane H+-ATPase. J. Biol. Chem. 272, 4795–4803 (1997)
Nishi, T., Kawasaki-Nishi, S. & Forgac, M. The first putative transmembrane segment of subunit c′′ (Vma16p) of the yeast V-ATPase is not necessary for function. J. Biol. Chem. 278, 5821–5827 (2003)
Matthies, D. et al. High-resolution structure and mechanism of an F/V-hybrid rotor ring in a Na+-coupled ATP synthase. Nature Commun. 5, 5286 (2014)
Nishi, T., Kawasaki-Nishi, S. & Forgac, M. Expression and localization of the mouse homologue of the yeast V-ATPase 21-kDa subunit c′′ (Vma16p). J. Biol. Chem. 276, 34122–34130 (2001)
Powell, B., Graham, L. A. & Stevens, T. H. Molecular characterization of the yeast vacuolar H+-ATPase proton pore. J. Biol. Chem. 275, 23654–23660 (2000)
Finnigan, G. C., Hanson-Smith, V., Stevens, T. H. & Thornton, J. W. Evolution of increased complexity in a molecular machine. Nature 481, 360–364 (2012)
Stock, D., Leslie, A. G. & Walker, J. E. Molecular architecture of the rotary motor in ATP synthase. Science 286, 1700–1705 (1999)
Nicholls, D. G. & Ferguson, S. J. Bioenergetics 3rd edn, Ch. 3 (Academic, 2002)
Bueler, S. A. & Rubinstein, J. L. Vma9p need not be associated with the yeast V-ATPase for fully-coupled proton pumping activity in vitro. Biochemistry 54, 853–858 (2015)
Toei, M., Toei, S. & Forgac, M. Definition of membrane topology and identification of residues important for transport in subunit a of the vacuolar ATPase. J. Biol. Chem. 286, 35176–35186 (2011)
Junge, W. & Nelson, N. Structural biology. Nature’s rotary electromotors. Science 308, 642–644 (2005)
Lau, W. C. Y. & Rubinstein, J. L. Subnanometre-resolution structure of the intact Thermus thermophilus H+-driven ATP synthase. Nature 481, 214–218 (2012)
Abrahams, J. P., Leslie, A. G., Lutter, R. & Walker, J. E. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature 370, 621–628 (1994)
Cingolani, G. & Duncan, T. M. Structure of the ATP synthase catalytic complex (F(1)) from Escherichia coli in an autoinhibited conformation. Nature Struct. Mol. Biol. 18, 701–707 (2011)
Numoto, N., Hasegawa, Y., Takeda, K. & Miki, K. Inter-subunit interaction and quaternary rearrangement defined by the central stalk of prokaryotic V1-ATPase. EMBO Rep. 10, 1228–1234 (2009)
Kabaleeswaran, V. et al. Asymmetric structure of the yeast F1 ATPase in the absence of bound nucleotides. J. Biol. Chem. 284, 10546–10551 (2009)
Wächter, A. et al. Two rotary motors in F-ATP synthase are elastically coupled by a flexible rotor and a stiff stator stalk. Proc. Natl Acad. Sci. USA 108, 3924–3929 (2011)
Marr, C. R., Benlekbir, S. & Rubinstein, J. L. Fabrication of carbon films with approximately 500 nm holes for cryo-EM with a direct detector device. J. Struct. Biol. 185, 42–47 (2014)
Rubinstein, J. L. & Brubaker, M. A. Alignment of cryo-EM movies of individual particles by optimization of image translations. ArXiv 1409, 1–11 (2014)
Mindell, J. A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003)
Zhao, J., Brubaker, M. A. & Rubinstein, J. L. TMaCS: a hybrid template matching and classification system for partially-automated particle selection. J. Struct. Biol. 181, 234–242 (2013)
Zhao, J., Brubaker, M. A., Benlekbir, S. & Rubinstein, J. L. Description and comparison of algorithms for correcting anisotropic magnification in cryo-EM images. ArXiv 1501, 1–10 (2015)
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012)
Scheres, S. H. W. et al. Disentangling conformational states of macromolecules in 3D-EM through likelihood optimization. Nature Methods 4, 27–29 (2007)
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nature Methods 11, 63–65 (2014)
Loken, C. et al. SciNet: lessons learned from building a power-efficient top-20 system and data centre. J. Phys. Conf. Ser. 256, 012026 (2010)
Goddard, T. D., Huang, C. C. & Ferrin, T. E. Visualizing density maps with UCSF Chimera. J. Struct. Biol. 157, 281–287 (2007)
Pintilie, G. D., Zhang, J., Goddard, T. D., Chiu, W. & Gossard, D. C. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J. Struct Biol. 170, 427–438 (2010)
Kelley, L. A. & Sternberg, M. J. Protein structure prediction on the Web: a case study using the Phyre server. Nature Protocols 4, 363–371 (2009)
Söding, J., Biegert, A. & Lupas, A. N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, 244–248 (2005)
Sali, A. & Blundell, T. L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 (1993)
Drory, O., Frolow, F. & Nelson, N. Crystal structure of yeast V-ATPase subunit C reveals its stator function. EMBO Rep. 5, 1148–1152 (2004)
Sagermann, M., Stevens, T. H. & Matthews, B. W. Crystal structure of the regulatory subunit H of the V-type ATPase of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA 98, 7134–7139 (2001)
Balakrishna, A. M., Basak, S., Manimekalai, M. S. S. & Gruber, G. Crystal structure of subunits D and F in complex give insight into energy transmission of the eukaryotic V-ATPase from Saccharomyces cerevisiae. J. Biol. Chem. 290, 3183–3196 (2015)
Oot, R. A., Huang, L. S., Berry, E. A. & Wilkens, S. Crystal structure of the yeast vacuolar ATPase heterotrimeric EGC(head) peripheral stalk complex. Structure 20, 1881–1892 (2012)
Iwata, M. et al. Crystal structure of a central stalk subunit C and reversible association/dissociation of vacuole-type ATPase. Proc. Natl Acad. Sci. USA 101, 59–64 (2004)
Srinivasan, S., Vyas, N. K., Baker, M. L. & Quiocho, F. A. Crystal structure of the cytoplasmic N-terminal domain of subunit I, a homolog of subunit a, of V-ATPase. J. Mol. Biol. 412, 14–21 (2011)
Murata, T., Yamato, I., Kakinuma, Y., Leslie, A. G. & Walker, J. E. Structure of the rotor of the V-Type Na+-ATPase from Enterococcus hirae. Science 308, 654–659 (2005)
Chen, S. et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy 135, 24–35 (2013)
Acknowledgements
We thank P. Rosenthal, R. Henderson, V. Kanelis, and L. Kay for comments on the manuscript. J.Z. was supported by a Doctoral Postgraduate Scholarship from the Natural Sciences and Engineering Research Council of Canada and a Mary Gertrude l’Anson Scholarship. J.L.R. is the Canada Research Chair in Electron Cryomicroscopy. This work was supported by operating grant MOP 81294 from the Canadian Institutes of Health Research.
Author information
Authors and Affiliations
Contributions
S.B. and J.L.R. initiated the project. J.Z. and S.B. collected images and performed pre-processing steps. J.Z. performed the image analysis. J.Z. and J.L.R. interpreted the data, prepared figures, and wrote the manuscript. J.L.R. and J.Z. contributed new computer algorithms used in image analysis.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 V-ATPase subunits and rotation.
a, The V-ATPase from S. cerevisiae consists of subunits A3B3CDE3FG3Hacxc′yc′′zde, where x, y, and z denote unknown stoichiometries. Subunits with upper-case letter names correspond to components of the soluble V1 region while lower-case names denote components of the membrane-bound VO region. The e-subunit is not found in the detergent-solubilized S. cerevisiae V-ATPase. During rotary catalysis, ATP hydrolysis drives rotation of the rotor, consisting of subunits DFcxc′yc′′zd (outlined in black), which rotates relative to the rest of the enzyme. Upper inset, the three different nucleotide-binding sites of the V1 region can be found in three different conformations: ‘tight’ (where ATP is expected to be bound), ‘loose’ (where ADP is expected to be bound), and ‘open’ (where no nucleotide is bound). Lower inset, rotation of the cxc′yc′′z-ring against the a-subunit leads to proton translocation from the cytoplasmic side of the membrane to the luminal side of the membrane. Proton translocation occurs via two half-channels through the membrane. b, V-ATPase activity is regulated by reversible dissociation where the V1 region separates from the VO region. The H-subunit inhibits ATP hydrolysis in the dissociated V1 region. Proton translocation in the dissociated VO region is blocked by an unknown mechanism.
Extended Data Figure 2 Data collection.
a, A representative micrograph; examples of V-ATPase particle images are shown circled in red. These particle images were selected from the 200 candidate particle images identified automatically from template matching. b, Tracking of particle and other image feature trajectories with the alignparts_lmbfgs algorithm31. Trajectories are exaggerated by a factor of 5 to allow visualization.
Extended Data Figure 3 Three-dimensional maps from rotational states.
a, Surface rendered views of the three three-dimensional maps are shown. Scale bars, 25 Å. b, Fourier shell correlation (FSC) curves after a ‘gold standard’ refinement of the three maps are shown. The resolutions measured from these curves at a Fourier shell correlation of 0.143 are the same as the resolutions measured after correcting for masking effects by high-resolution noise-substitution calculations51. c, Local resolution estimation shows that features in the V1 region are better resolved than in the VO region.
Extended Data Figure 4 Map segmentation and molecular dynamics flexible fitting.
Different subunits are shown fitted into their corresponding map densities in rotational states 1, 2, and 3, including AB pair 3 (a), the N-terminal domain of subunit a (b), the central rotor DFd subcomplex (c), subunit C (d), and peripheral stalk 1 (e). Scale bar, 25 Å.
Extended Data Figure 5 C-terminal domain of the a-subunit.
a, The membrane-bound C-terminal domain of the a-subunit appears similar in all three rotational states. b, The density from the C-terminal domain of the Thermus thermophilus subunit I, equivalent of the a-subunit, at 9.7 Å resolution3 is consistent with the structure of the a-subunit from S. cerevisiae (left). However, the transmembrane α-helical densities identified previously in that map (right) are not entirely consistent with the current maps.
Extended Data Figure 6 Flexibility in V-ATPase subunits.
a–c, Each AB pair in the A3B3 hexamer goes through ‘open’, ‘loose’, and ‘tight’ conformations as the enzyme passes between the three rotational states. d–f, Overlay of all three open, all three loose, and all three tight structures shows that the conformations are nearly the same for each AB pair. g–i, Each of the three EG peripheral stalk structures undergoes similar bending motions between the three rotational states. Scale bar, 25 Å.
Supplementary information
Cross sections through the three maps, each showing a different rotational state of the V-ATPase.
Cross sections through the three maps, each showing a different rotational state of the V-ATPase. (MOV 12001 kb)
Interpolation between the three observed rotational states of the V-ATPase
Interpolation between the three observed rotational states of the V-ATPase. (MOV 32484 kb)
Exploded view of subunits when interpolating between the three observed rotational states of the V-ATPase
Exploded view of subunits when interpolating between the three observed rotational states of the V-ATPase. (MOV 30321 kb)
Rights and permissions
About this article
Cite this article
Zhao, J., Benlekbir, S. & Rubinstein, J. Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase. Nature 521, 241–245 (2015). https://doi.org/10.1038/nature14365
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14365