Abstract
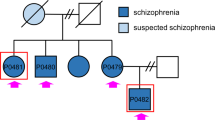
Dysregulated neurodevelopment with altered structural and functional connectivity is believed to underlie many neuropsychiatric disorders1, and ‘a disease of synapses’ is the major hypothesis for the biological basis of schizophrenia2. Although this hypothesis has gained indirect support from human post-mortem brain analyses2,3,4 and genetic studies5,6,7,8,9,10, little is known about the pathophysiology of synapses in patient neurons and how susceptibility genes for mental disorders could lead to synaptic deficits in humans. Genetics of most psychiatric disorders are extremely complex due to multiple susceptibility variants with low penetrance and variable phenotypes11. Rare, multiply affected, large families in which a single genetic locus is probably responsible for conferring susceptibility have proven invaluable for the study of complex disorders. Here we generated induced pluripotent stem (iPS) cells from four members of a family in which a frameshift mutation of disrupted in schizophrenia 1 (DISC1) co-segregated with major psychiatric disorders12 and we further produced different isogenic iPS cell lines via gene editing. We showed that mutant DISC1 causes synaptic vesicle release deficits in iPS-cell-derived forebrain neurons. Mutant DISC1 depletes wild-type DISC1 protein and, furthermore, dysregulates expression of many genes related to synapses and psychiatric disorders in human forebrain neurons. Our study reveals that a psychiatric disorder relevant mutation causes synapse deficits and transcriptional dysregulation in human neurons and our findings provide new insight into the molecular and synaptic etiopathology of psychiatric disorders.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Weinberger, D. R. Implications of normal brain development for the pathogenesis of schizophrenia. Arch. Gen. Psychiatry 44, 660–669 (1987)
Mirnics, K., Middleton, F. A., Lewis, D. A. & Levitt, P. Analysis of complex brain disorders with gene expression microarrays: schizophrenia as a disease of the synapse. Trends Neurosci. 24, 479–486 (2001)
Johnson, R. D., Oliver, P. L. & Davies, K. E. SNARE proteins and schizophrenia: linking synaptic and neurodevelopmental hypotheses. Acta Biochim. Pol. 55, 619–628 (2008)
Honer, W. G. & Young, C. E. Presynaptic proteins and schizophrenia. Int. Rev. Neurobiol. 59, 175–199 (2004)
Gulsuner, S. et al. Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell 154, 518–529 (2013)
Kenny, E. M. et al. Excess of rare novel loss-of-function variants in synaptic genes in schizophrenia and autism spectrum disorders. Mol. Psychiatry 19, 872–879 (2014)
Malhotra, D. et al. High frequencies of de novo CNVs in bipolar disorder and schizophrenia. Neuron 72, 951–963 (2011)
Purcell, S. M. et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature 506, 185–190 (2014)
Fromer, M. et al. De novo mutations in schizophrenia implicate synaptic networks. Nature 506, 179–184 (2014)
Lips, E. S. et al. Functional gene group analysis identifies synaptic gene groups as risk factor for schizophrenia. Mol. Psychiatry 17, 996–1006 (2012)
Sullivan, P. F., Daly, M. J. & O’Donovan, M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nature Rev. Genet. 13, 537–551 (2012)
Sachs, N. A. et al. A frameshift mutation in Disrupted in Schizophrenia 1 in an American family with schizophrenia and schizoaffective disorder. Mol. Psychiatry 10, 758–764 (2005)
Thomson, P. A. et al. DISC1 genetics, biology and psychiatric illness. Front. Biol. 8, 1–31 (2013)
Duan, X. et al. Disrupted-In-Schizophrenia 1 regulates integration of newly generated neurons in the adult brain. Cell 130, 1146–1158 (2007)
Christian, K., Song, H. & Ming, G. Application of reprogrammed patient cells to investigate the etiology of neurological and psychiatric disorders. Front. Biol. 7, 179–188 (2012)
Chiang, C. H. et al. Integration-free induced pluripotent stem cells derived from schizophrenia patients with a DISC1 mutation. Mol. Psychiatry 16, 358–360 (2011)
Kuroda, K. et al. Behavioral alterations associated with targeted disruption of exons 2 and 3 of the Disc1 gene in the mouse. Hum. Mol. Genet. 20, 4666–4683 (2011)
Leliveld, S. R. et al. Insolubility of disrupted-in-schizophrenia 1 disrupts oligomer-dependent interactions with nuclear distribution element 1 and is associated with sporadic mental disease. J. Neurosci. 28, 3839–3845 (2008)
Custer, K. L., Austin, N. S., Sullivan, J. M. & Bajjalieh, S. M. Synaptic vesicle protein 2 enhances release probability at quiescent synapses. J. Neurosci. 26, 1303–1313 (2006)
Chang, W. P. & Südhof, T. C. SV2 renders primed synaptic vesicles competent for Ca2+-induced exocytosis. J. Neurosci. 29, 883–897 (2009)
Marchetto, M. C. et al. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell 143, 527–539 (2010)
Camargo, L. M. et al. Disrupted in Schizophrenia 1 Interactome: evidence for the close connectivity of risk genes and a potential synaptic basis for schizophrenia. Mol. Psychiatry 12, 74–86 (2007)
Hackett, J. T., Cochran, S. L., Greenfield, L. J., Jr, Brosius, D. C. & Ueda, T. Synapsin I injected presynaptically into goldfish mauthner axons reduces quantal synaptic transmission. J. Neurophysiol. 63, 701–706 (1990)
Rosahl, T. W. et al. Short-term synaptic plasticity is altered in mice lacking synapsin I. Cell 75, 661–670 (1993)
Flavell, S. W. et al. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science 311, 1008–1012 (2006)
Barbosa, A. C. et al. MEF2C, a transcription factor that facilitates learning and memory by negative regulation of synapse numbers and function. Proc. Natl Acad. Sci. USA 105, 9391–9396 (2008)
Wright, R., Rethelyi, J. M. & Gage, F. H. Enhancing induced pluripotent stem cell models of schizophrenia. JAMA Psychiatry 71, 334–335 (2014)
Brennand, K. J. et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature 473, 221–225 (2011)
Yu, D. X. et al. Modeling hippocampal neurogenesis using human pluripotent stem cells. Stem Cell Reports 2, 295–310 (2014)
Brennand, K. et al. Phenotypic differences in hiPS cells NPCs derived from patients with schizophrenia. Mol. Psychiatry http://dx.doi.org/10.1038/mp.2014.22 (2014)
Juopperi, T. A. et al. Astrocytes generated from patient induced pluripotent stem cells recapitulate features of Huntington’s disease patient cells. Mol. Brain 5, 17 (2012)
Ma, D. K. et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science 323, 1074–1077 (2009)
Cermak, T. et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 39, e82 (2011)
Song, H. J., Stevens, C. F. & Gage, F. H. Neural stem cells from adult hippocampus develop essential properties of functional CNS neurons. Nature Neurosci. 5, 438–445 (2002)
Ho, S. Y. et al. NeurphologyJ: an automatic neuronal morphology quantification method and its application in pharmacological discovery. BMC Bioinformatics 12, 230 (2011)
Trapnell, C. et al. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnol. 28, 511–515 (2010)
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nature Methods 9, 357–359 (2012)
Wang, L., Wang, S. & Li, W. RSeQC: quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185 (2012)
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010)
Wang, J., Duncan, D., Shi, Z. & Zhang, B. WEB-based gene set analysis toolkit (WebGestalt): update 2013. Nucleic Acids Res. 41, W77–W83 (2013)
Acknowledgements
We thank members of Ming and Song laboratories for discussion, and Q. Hussaini, Y. Cai and L. Liu for technical support. This work was supported by grants from the NIH (MH087874, NS047344), IMHRO, SFARI, NARSAD, and MSCRF to H.S.; from MSCRF, NARSAD and the NIH (NS048271) to G.-l.M.; from Dr. Miriam and Sheldon G. Adelson Medical Research Foundation to G.-l.M. and K.S.K.; from the NIH (AG045656) to G.C.; from MSCRF and NARSAD to K.M.C.; by postdoctoral fellowships from MSCRF to Z.W., Y.S., N.S.K., and G.M.; and by a predoctoral fellowship from the NIH (MH102978) to H.N.N.
Author information
Authors and Affiliations
Contributions
Z.W. led and was involved in every aspect of the project. H.N.N. generated isogenic iPS cell lines. Z.G. and G.C. performed electrophysiology analyses. M.A.L., E.G. and K.S.K. performed RNA-seq analyses. X.W., Y.S., N.-S.K., K.-J.Y., J.S., C.Z., G.M., D.N., H.Y., C.-H.C. and K.M.C. helped with data collection. K.K. provided DISC1 antibodies. N.Y., C.A.R. and R.L.M. obtained original skin biopsies from pedigree H.J.Z. and L.C. helped with TALEN design. G.-l.M., H.S. and Z.W. designed the project and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Basic characterization of iPS cell lines.
a–c, Sample confocal images of immunostaining of pluripotency-associated markers for different iPS cell lines (a, scale bar, 50 μm) and sample images of karyotyping (b). Also shown is sample bisulphite-sequencing analysis of promoter regions of pluripotency genes NANOG and OCT4 (c). Each row represents one allele: closed circles represent methylated cytosine and open circles represent unmethylated cytosine. d, e, Pluripotency of iPS cell lines. Shown are sample images of cell types of three germ-layers in teratomas following transplantation to SCID mice (d, scale bar, 100 μm) and immunostaining for AFP (an endoderm marker), SMA (a mesoderm marker) or TUJ1 (an ectoderm/neuronal marker), upon in vitro differentiation of iPS cells (e, scale bar, 50 μm). f, Confirmation of the genotype of different iPS cell lines by Sanger sequencing. Shown are sample genomic DNA sequences around exon 12 and intron 12 of different iPS cell lines. Each line represents one allele. See Supplementary Table 1a for a summary of similar characterization for all iPS cell lines used in this study.
Extended Data Figure 2 Forebrain-specific neural differentiation of iPS cell lines.
a, Schematic diagram of the differentiation procedure. b, Sample confocal images of immunostaining for nestin and forebrain progenitor markers, EMX1, FOXG1, OTX2, and PAX6, and DAPI. Scale bar, 20 μm.
Extended Data Figure 3 Neuronal subtype differentiation of iPS cell lines.
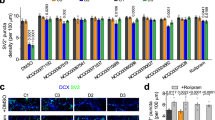
a, Expression of glutamatergic neuron marker α-CAMKII. Shown are sample confocal images of immunostaining of CAMKII and MAP2AB and quantification. Scale bar, 20 μm. Values represent mean ± s.e.m. n = 5 cultures. b, Expression of GABAergic neuron marker GAD67. Same as in a, except that GAD67 was examined. c, Expression of dopaminergic neuron marker tyrosine hydroxylase in cultures. Same as in a, except that tyrosine hydroxylase was examined in one iPS cell line each from five individuals.
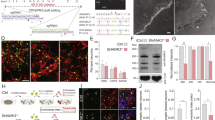
Extended Data Figure 4 Effect of the 4-bp deletion mutation of DISC1 on wild-type DISC1 at the protein level.
a, Schematic diagram of the DISC1 locus harbouring the frameshift 4-bp deletion mutation. Also shown are predicated protein sequences at the C terminus of wDISC1 and mDISC1. b, Quantification of DISC1 mRNA levels from qPCR analysis of exon 2. Data were normalized to that of C3-1 neurons. Values represent mean ± s.e.m., n = 3. c, Sample western blot images of co-immunoprecipitation analyses of differentially tagged wDISC1 and mDISC1 upon co-expression in HEK293 cells. d, Dose-dependent depletion of soluble wDISC1 by mDISC1 upon co-expression in HEK293 cells. Shown are sample western blot images and quantification. Data were normalized to that of the 2:2 ratio condition for each experiment. Values represent mean ± s.e.m. (n = 3). e, Increased ubiquitination of wDISC1 upon mDISC1co-expression. Expression plasmids for V5-tagged ubiquitin and HA-tagged wDISC1 were co-transfected with or without Flag-tagged mDISC1 into HEK293 cells. Samples were prepared with lysate buffer containing SDS to dissociate the protein complex, and then wDISC1 was immunoprecipitated with anti-HA antibodies, followed by western blot analysis using anti-V5 antibodies. Note markedly increased covalent-bound ubiquitin for wDISC1 upon mDISC1 co-expression.
Extended Data Figure 5 Morphological development of forebrain neurons in culture.
Shown are summaries of soma size and total dendritic length of forebrain neurons derived from two iPS cell lines from each individual at 1 to 4 weeks after neuronal differentiation. Numbers associated with the bars indicate total numbers of neurons examined. Values represent mean ± s.e.m. n = 5 cultures; ANOVA analysis.
Extended Data Figure 6 I-V characteristics of forebrain neurons derived from different iPS cell lines.
Shown are summaries of recordings from forebrain neurons derived from 4 iPS cell lines in co-culture with astrocytes for 1, 2 or 4 weeks. Values represent mean ± s.e.m., n = 6–15 cells for each condition.
Extended Data Figure 7 Basic characterization of isogenic iPS cell lines.
a, b, Sample images of immunostaining of pluripotency-associated markers for different isogenic iPS cell lines (a; scale bars, 50 μm) and sample images for karyotyping (b). See Supplementary Table 1a for a summary.
Extended Data Figure 8 Validations of differential gene and protein expression in forebrain neurons from different isogenic iPS cell lines.
a, Heat-map of expression profile of 500 genes. b, Dot plot of gene expression analysis of a selected group of 23 genes from RNA-seq and qRT–PCR analyses of independent samples of C3-1 and D2-1 neurons. Data represent mean values (n = 3). c, Quantitative mRNA analysis of a selected group of synapse-related genes in forebrain neurons from different isogenic lines. Data from RNA-seq analysis (n = 3 samples each) are also shown for comparison. Values represent mean ± s.e.m. (n = 3; *P < 0.01; ANOVA). The same data are summarized in a heat-map illustration shown in Fig. 4d. d, Quantitative analysis of protein expression based on western blot analysis. Values represent mean ± s.e.m. (n = 3; *P < 0.01; ANOVA). The same data are summarized in a heat-map illustration shown in Fig. 4e.
Supplementary information
Supplementary Table 1
Summary of iPSC lines and reagents used in the current study. a, Summary of characterization of all iPSC lines used. b, Summary of information for antibodies used. c, List of primer sequences. (XLS 51 kb)
Supplementary Table 2
Summary of RNA-seq analysis of 4 week-old forebrain neurons from D2-1, D3-2 and C3-1 iPSC lines. a, RNA-seq read information. b, List of common up-regulated genes in DISC1 mutant D2-1 and D3-2 forebrain neurons compared to control C3-1 neurons. c, List of common down-regulated genes in DISC1 mutant D2-1 and D3-2 forebrain neurons compared to control C3-1 neurons; d, List of differentially expressed genes related to synapses; e, List of differentially expressed genes related to mental disorders. (XLS 522 kb)
Rights and permissions
About this article
Cite this article
Wen, Z., Nguyen, H., Guo, Z. et al. Synaptic dysregulation in a human iPS cell model of mental disorders. Nature 515, 414–418 (2014). https://doi.org/10.1038/nature13716
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13716