Abstract
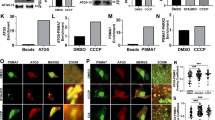
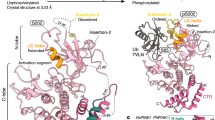
The PARKIN ubiquitin ligase (also known as PARK2) and its regulatory kinase PINK1 (also known as PARK6), often mutated in familial early-onset Parkinson’s disease, have central roles in mitochondrial homeostasis and mitophagy1,2,3. Whereas PARKIN is recruited to the mitochondrial outer membrane (MOM) upon depolarization via PINK1 action and can ubiquitylate porin, mitofusin and Miro proteins on the MOM1,4,5,6,7,8,9,10,11, the full repertoire of PARKIN substrates—the PARKIN-dependent ubiquitylome—remains poorly defined. Here we use quantitative diGly capture proteomics (diGly)12,13 to elucidate the ubiquitylation site specificity and topology of PARKIN-dependent target modification in response to mitochondrial depolarization. Hundreds of dynamically regulated ubiquitylation sites in dozens of proteins were identified, with strong enrichment for MOM proteins, indicating that PARKIN dramatically alters the ubiquitylation status of the mitochondrial proteome. Using complementary interaction proteomics, we found depolarization-dependent PARKIN association with numerous MOM targets, autophagy receptors, and the proteasome. Mutation of the PARKIN active site residue C431, which has been found mutated in Parkinson’s disease patients, largely disrupts these associations. Structural and topological analysis revealed extensive conservation of PARKIN-dependent ubiquitylation sites on cytoplasmic domains in vertebrate and Drosophila melanogaster MOM proteins. These studies provide a resource for understanding how the PINK1–PARKIN pathway re-sculpts the proteome to support mitochondrial homeostasis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
17 April 2013
Two gene names were corrected (PDCD6IP and IER3IP1) and an indicated site on CLTC and MAOB was removed.
References
Narendra, D., Walker, J. E. & Youle, R. Mitochondrial quality control mediated by PINK1 and Parkin: links to parkinsonism. Cold Spring Harb. Perspect. Biol. 4, a011338 (2012)
Youle, R. J. & Narendra, D. P. Mechanisms of mitophagy. Nature Rev. Mol. Cell Biol. 12, 9–14 (2011)
Dawson, T. M. & Dawson, V. L. The role of parkin in familial and sporadic Parkinson’s disease. Mov. Disord. 25 (Suppl 1). S32–S39 (2010)
Chan, N. C. et al. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum. Mol. Genet. 20, 1726–1737 (2011)
Glauser, L., Sonnay, S., Stafa, K. & Moore, D. J. Parkin promotes the ubiquitination and degradation of the mitochondrial fusion factor mitofusin 1. J. Neurochem. 118, 636–645 (2011)
Ziviani, E., Tao, R. N. & Whitworth, A. J. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc. Natl Acad. Sci. USA 107, 5018–5023 (2010)
Tanaka, A. et al. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J. Cell Biol. 191, 1367–1380 (2010)
Poole, A. C., Thomas, R. E., Yu, S., Vincow, E. S. & Pallanck, L. The mitochondrial fusion-promoting factor mitofusin is a substrate of the PINK1/parkin pathway. PLoS ONE 5, e10054 (2010)
Sun, Y., Vashisht, A. A., Tchieu, J., Wohlschlegel, J. A. & Dreier, L. VDACs recruit Parkin to defective mitochondria to promote mitochondrial autophagy. J. Biol. Chem. 287, 40652–40660 (2012)
Wang, X. et al. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell 147, 893–906 (2011)
Liu, S. et al. Parkinson’s disease-associated kinase PINK1 regulates Miro protein level and axonal transport of mitochondria. PLoS Genet. 8, e1002537 (2012)
Kim, W. et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 (2011)
Wagner, S. A. et al. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell Proteomics 10, M111.013284 (2011)
Wenzel, D. M., Lissounov, A., Brzovic, P. S. & Klevit, R. E. UbcH7-reactivity profile reveals Parkin and HHARI to be RING/HECT hybrids. Nature 474, 105–108 (2011)
Joselin, A. P. et al. ROS-dependent regulation of Parkin and DJ-1 localization during oxidative stress in neurons. Hum. Mol. Genet. 21, 4888–4903 (2012)
Narendra, D., Tanaka, A., Suen, D. F. & Youle, R. J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183, 795–803 (2008)
Lazarou, M., Jin, S. M., Kane, L. A. & Youle, R. J. Role of PINK1 binding to the TOM complex and alternate intracellular membranes in recruitment and activation of the E3 ligase Parkin. Dev. Cell 22, 320–333 (2012)
Kondapalli, C. et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2, 120080 (2012)
Sha, D., Chin, L. S. & Li, L. Phosphorylation of parkin by Parkinson disease-linked kinase PINK1 activates parkin E3 ligase function and NF-κB signaling. Hum. Mol. Genet. 19, 352–363 (2010)
Lazarou, M. et al. PINK1 drives Parkin self-association and HECT-like E3 activity upstream of mitochondrial binding. J. Cell Biol. 200, 163–172 (2013)
Geisler, S. et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nature Cell Biol. 12, 119–131 (2010)
Sowa, M. E., Bennett, E. J., Gygi, S. P. & Harper, J. W. Defining the human deubiquitinating enzyme interaction landscape. Cell 138, 389–403 (2009)
Chaugule, V. K. et al. Autoregulation of Parkin activity through its ubiquitin-like domain. EMBO J. 30, 2853–2867 (2011)
Narendra, D., Kane, L. A., Hauser, D. N., Fearnley, I. M. & Youle, R. J. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy 6, 1090–1106 (2010)
Onoue, K. et al. Fis1 acts as mitochondrial recruitment factor for TBC1D15 that is involved in regulation of mitochondrial morphology. J. Cell Sci.. http://dx.doi.org/10.1242/jcs.111211 (2012)
von Muhlinen, N. et al. LC3C, bound selectively by a noncanonical LIR motif in NDP52, is required for antibacterial autophagy. Mol. Cell 48, 329–342 (2012)
Huang, P., Galloway, C. A. & Yoon, Y. Control of mitochondrial morphology through differential interactions of mitochondrial fusion and fission proteins. PLoS ONE 6, e20655 (2011)
Villén, J. & Gygi, S. P. The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nature Protocols 3, 1630–1638 (2008)
Haas, W. et al. Optimization and use of peptide mass measurement accuracy in shotgun proteomics. Mol. Cell. Proteomics 5, 1326–1337 (2006)
Huttlin, E. L. et al. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189 (2010)
Eng, J. K., McCormack, A. L. & Yates, J. R., III An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 (1994)
Beausoleil, S. A., Villen, J., Gerber, S. A., Rush, J. & Gygi, S. P. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nature Biotechnol. 24, 1285–1292 (2006)
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols 4, 44–57 (2009)
Acknowledgements
We thank W. Kim for LC-MS and for development of QdiGly profiling, R. Kunz for peptide purification, J. Lydeard, S. Hayes and A. White for proteomics, M. Comb and S. Beausoleil (Cell Signaling Technologies) for antibodies, Nikon Imaging Center (Harvard Medical School) for microscopy, and D. Finley and B. Schulman for discussions. Supported by NIH grants GM070565 and GM095567 to J.W.H., GM067945 to S.P.G., CA139885 to M.R., and the Michael J. Fox Foundation for Parkinson’s Research to J.W.H.
Author information
Authors and Affiliations
Contributions
S.A.S. and J.W.H. conceived the experiments. S.A.S. performed QdiGly profiling, biochemical, and interaction experiments and analysis. S.A.S., M.R. and V.G.-P. performed cell biological experiments. M.E.S. designed site visualization software. E.L.H. and S.P.G. provided proteomics software and analysis. S.A.S. and J.W.H. wrote the manuscript. All authors assisted in editing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-7, Supplementary Text and Supplementary References. (PDF 11288 kb)
Supplementary Table 1
This file contains experimental parameters of QdiGLY proteomic experiments reported in this study, It shows the experiment numbers in relation to cell lines used, treatments employed, and the number of sequential immunoprecipitations. (XLSX 44 kb)
Supplementary Table 2
This file contains a complete list of all proteins and sites identified and quantified from all experiments from this study, as well as Tier1, 2, and 3 and Class1 and Class 2 site lists. (XLSX 15491 kb)
Supplementary Table 3
This file contains proteomic analysis of HA-FLAG-PARKIN associated proteins in 293T cells in response to depolarization using CompPASS. It shows all the WDN-scores, Z-scores, and APSMs for the PARKIN immunoprecipitation data from 293T cells. (XLSX 5435 kb)
Supplementary Table 4
This file contains proteomic analysis of HA-FLAG-PARKIN associated proteins in HeLa cells in response to depolarization using CompPASS. It shows all the WDN-scores, Z-scores, and APSMs for the PARKIN immunoprecipitation data from HeLa cells. (XLSX 212 kb)
Supplementary Table 5
This file shows conservation and structural analysis of selected candidate PARKIN substrates. This file contains Protein DataBase (PDB) identifiers, the identity of ubiquitylation sites that change in response to depolarization, and the conservation of sites in M. musculus, D. rerio, and D. melanogaster. (XLSX 40 kb)
Supplementary Table 6
This file contains quantification of the PARKIN-mitochondrial overlap for the imaging experiments in Supplementary Figure 6. (XLSX 12 kb)
Rights and permissions
About this article
Cite this article
Sarraf, S., Raman, M., Guarani-Pereira, V. et al. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature 496, 372–376 (2013). https://doi.org/10.1038/nature12043
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12043