Abstract
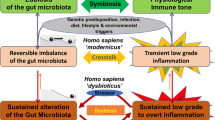
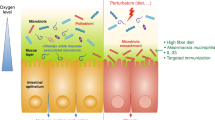
Trillions of microbes inhabit the human intestine, forming a complex ecological community that influences normal physiology and susceptibility to disease through its collective metabolic activities and host interactions. Understanding the factors that underlie changes in the composition and function of the gut microbiota will aid in the design of therapies that target it. This goal is formidable. The gut microbiota is immensely diverse, varies between individuals and can fluctuate over time — especially during disease and early development. Viewing the microbiota from an ecological perspective could provide insight into how to promote health by targeting this microbial community in clinical treatments.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Candela, M. et al. Interaction of probiotic Lactobacillus and Bifidobacterium strains with human intestinal epithelial cells: adhesion properties, competition against enteropathogens and modulation of IL-8 production. Int. J. Food Microbiol. 125, 286–292 (2008).
Fukuda, S. et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 469, 543–547 (2011).
Sonnenburg, J. L. et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 307, 1955–1959 (2005).
Yatsunenko, T. et al. Human gut microbiome viewed across age and geography. Nature 486, 222–227 (2012).
Olszak, T. et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science 336, 489–493 (2012).
Ley, R. E., Turnbaugh, P. J., Klein, S. & Gordon, J. I. Microbial ecology: human gut microbes associated with obesity. Nature 444, 1022–1023 (2006).
Turnbaugh, P. J., Backhed, F., Fulton, L. & Gordon, J. I. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 3, 213–223 (2008).
Kau, A. L., Ahern, P. P., Griffin, N. W., Goodman, A. L. & Gordon, J. I. Human nutrition, the gut microbiome and the immune system. Nature 474, 327–336 (2011).
Dicksved, J. et al. Molecular analysis of the gut microbiota of identical twins with Crohn's disease. ISME J. 2, 716–727 (2008).
Frank, D. N. et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl Acad. Sci. USA 104, 13780–13785 (2007).
Gonzalez, A. et al. The mind–body–microbial continuum. Dialogues Clin. Neurosci. 13, 55–62 (2011).
Lupton, J. R. Microbial degradation products influence colon cancer risk: the butyrate controversy. J. Nutr. 134, 479–482 (2004).
Qin, J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010).
The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
Borenstein, E., Kupiec, M., Feldman, M. W. & Ruppin, E. Large-scale reconstruction and phylogenetic analysis of metabolic environments. Proc. Natl Acad. Sci. USA 105, 14482–14487 (2008).
Freilich, S. et al. Metabolic-network-driven analysis of bacterial ecological strategies. Genome Biol. 10, R61 (2009).
Claesson, M. J. et al. Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS ONE 4, e6669 (2009).
Eckburg, P. B. et al. Diversity of the human intestinal microbial flora. Science 308, 1635–1638 (2005).
Reyes, A. et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 466, 334–338 (2010).
Turnbaugh, P. J. et al. A core gut microbiome in obese and lean twins. Nature 457, 480–484 (2009).
Biagi, E. et al. Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS ONE 5, e10667 (2010).
Nelson, K. E. et al. A catalog of reference genomes from the human microbiome. Science 328, 994–999 (2010).
Verberkmoes, N. C. et al. Shotgun metaproteomics of the human distal gut microbiota. ISME J. 3, 179–189 (2009).
Jansson, J. et al. Metabolomics reveals metabolic biomarkers of Crohn's disease. PLoS ONE 4, e6386 (2009).
Muegge, B. D. et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332, 970–974 (2011).
Koenig, J. E. et al. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl Acad. Sci. USA 108, 4578–4585 (2011).
Palmer, C., Bik, E. M., DiGiulio, D. B., Relman, D. A. & Brown, P. O. Development of the human infant intestinal microbiota. PLoS Biol. 5, e177 (2007).
Dominguez-Bello, M. G. et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl Acad. Sci. USA 107, 11971–11975 (2010).
Kozyrskyj, A. L., Bahreinian, S. & Azad, M. B. Early life exposures: impact on asthma and allergic disease. Curr. Opin. Allergy Clin. Immunol. 11, 400–406 (2011).
De Filippo, C. et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl Acad. Sci. USA 107, 14691–14696 (2010).
Arumugam, M. et al. Enterotypes of the human gut microbiome. Nature 473, 174–180 (2011). This paper reports that there is an association between co-occurring microbial groups, and that high Prevotella versus Bacteroides genus level abundance estimates are associated with major patterns of differentiation in the microbiota across people.
Wu, G. D. et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 334, 105–108 (2011). This study found a strong correlation between microbiota diversity and long-term diets as assessed using diet inventories.
Loftus, E. V. Jr. Clinical epidemiology of inflammatory bowel disease: incidence, prevalence, and environmental influences. Gastroenterology 126, 1504–1517 (2004).
Cann, H. M. et al. A human genome diversity cell line panel. Science 296, 261–262 (2002).
Bach, J. F. & Chatenoud, L. The hygiene hypothesis: an explanation for the increased frequency of insulin-dependent diabetes. Cold Spring Harb. Perspect. Med. 2, a007799 (2012).
Clayton, T. A., Baker, D., Lindon, J. C., Everett, J. R. & Nicholson, J. K. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc. Natl Acad. Sci. USA 106, 14728–14733 (2009).
Jackson, R. L., Greiwe, J. S. & Schwen, R. J. Emerging evidence of the health benefits of S-equol, an estrogen receptor beta agonist. Nutr. Rev. 69, 432–448 (2011).
Setchell, K. D. & Clerici, C. Equol: history, chemistry, and formation. J. Nutr. 140, 1355S–1362S (2010).
Caporaso, J. G. et al. Moving pictures of the human microbiome. Genome Biol. 12, R50 (2011).
Costello, E. K. et al. Bacterial community variation in human body habitats across space and time. Science 326, 1694–1697 (2009).
Dethlefsen, L. & Relman, D. A. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc. Natl Acad. Sci. USA 108, 4554–4561 (2011). This paper gives insight into the resilience of the human microbiota in the face of repeated disturbances, and the degree of baseline variation.
Jakobsson, H. E. et al. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS ONE 5, e9836 (2010).
Beisner, B. E., Haydon, D. T. & Cuddington, K. Alternative stable states in ecology. Front. Ecol. Environ. 1, 376–382 (2003).
Walker, B., Hollin, C. S., Carpenter, S. R. & Kinzig, A. Resilience, adaptability and transformability in social-ecological systems. Ecol. Soc. 9, http://www.ecologyandsociety.org/vol9/iss2/art5 (16 September, 2004).
Turnbaugh, P. J. et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031 (2006).
Sun, Y. et al. Advanced computational algorithms for microbial community analysis using massive 16S rRNA sequence data. Nucleic Acids Res. 38, e205 (2010).
Knights, D., Parfrey, L. W., Zaneveld, J., Lozupone, C. & Knight, R. Human-associated microbial signatures: examining their predictive value. Cell Host Microbe 10, 292–296 (2011).
Carroll, I. M. et al. Molecular analysis of the luminal- and mucosal-associated intestinal microbiota in diarrhea-predominant irritable bowel syndrome. Am. J. Physiol. Gastrointest. Liver Physiol. 301, G799–G807 (2011).
Chang, J. Y. et al. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J. Infect. Dis. 197, 435–438 (2008).
Young, V. B. & Schmidt, T. M. Antibiotic-associated diarrhea accompanied by large-scale alterations in the composition of the fecal microbiota. J. Clin. Microbiol. 42, 1203–1206 (2004).
Willing, B. P. et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 139, 1844–1854 (2010).
Swidsinski, A., Loening-Baucke, V. & Herber, A. Mucosal flora in Crohn's disease and ulcerative colitis — an overview. J. Physiol. Pharmacol. 60, 61–71 (2009).
Lozupone, C. et al. Identifying genomic and metabolic features that can underlie early successional and opportunistic lifestyles in human gut symbionts. Genome Res. http://dx.doi.org/10.1101/gr.138198.112 (4 June, 2012).
Libby, J. M., Donta, S. T. & Wilkins, T. D. Clostridium difficile toxin A in infants. J. Infect. Dis. 148, 606 (1983).
Yamamoto-Osaki, T., Kamiya, S., Sawamura, S., Kai, M. & Ozawa, A. Growth inhibition of Clostridium difficile by intestinal flora of infant faeces in continuous flow culture. J. Med. Microbiol. 40, 179–187 (1994).
Folke, C. et al. Regime shifts, resilience, and biodiversity in ecosystem management. Annu. Rev. Ecol. Evol. Syst. 35, 557–581 (2004).
Scheffer, M. et al. Floating plant dominance as a stable state. Proc. Natl Acad. Sci. USA 100, 4040–4045 (2003).
Hazen, T. C. et al. Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science 330, 204–208 (2010).
Valentine, D. L. et al. Dynamic autoinoculation and the microbial ecology of a deep water hydrocarbon irruption. Proc. Natl Acad. Sci. USA http://dx.doi.org/10.1073/pnas.1108820109 (10 January, 2012).
Jernberg, C., Lofmark, S., Edlund, C. & Jansson, J. K. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J. 1, 56–66 (2007).
van der Waaij, D., Berghuis, J. M. & Lekkerkerk, J. E. Colonization resistance of the digestive tract of mice during systemic antibiotic treatment. J. Hyg. (Lond.) 70, 605–610 (1972).
McNulty, N. P. et al. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci. Transl. Med. 3, 106ra106 (2011).
Manichanh, C. et al. Reshaping the gut microbiome with bacterial transplantation and antibiotic intake. Genome Res. 20, 1411–1419 (2010). This study indicates that the indigenous microbiota may be more plastic than previously thought. The observation that antibiotic pretreatment interfered with, rather than promoted, establishment of the donor community indicates that low species abundance or diversity alone cannot predict low colonization resistance.
Levine, J. M. & D'antonio, C. M. Elton revisited: a review of evidence linking diversity and invasibility. Oikos 87, 15–26 (1999).
Khoruts, A., Dicksved, J., Jansson, J. K. & Sadowsky, M. J. Changes in the composition of the human fecal microbiome after bacteriotherapy for recurrent Clostridium difficile-associated diarrhea. J. Clin. Gastroenterol. 44, 354–360 (2010).
Gough, E., Shaikh, H. & Manges, A. R. Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clin. Infect. Dis. 53, 994–1002 (2011).
Hautier, Y., Niklaus, P. A. & Hector, A. Competition for light causes plant biodiversity loss after eutrophication. Science 324, 636–638 (2009).
Elmqvist, T. et al. Response diversity, ecosystem change, and resilience. Front. Ecol. Environ. 1, 488–494 (2003).
Hansen, E. E. et al. Pan-genome of the dominant human gut-associated archaeon, Methanobrevibacter smithii, studied in twins. Proc. Natl Acad. Sci. USA 108, 4599–4606 (2011).
Flint, H. J., Duncan, S. H., Scott, K. P. & Louis, P. Interactions and competition within the microbial community of the human colon: links between diet and health. Environ. Microbiol. 9, 1101–1111 (2007).
Louis, P. et al. Restricted distribution of the butyrate kinase pathway among butyrate-producing bacteria from the human colon. J. Bacteriol. 186, 2099–2106 (2004).
Chaffron, S., Rehrauer, H., Pernthaler, J. & von Mering, C. A global network of coexisting microbes from environmental and whole-genome sequence data. Genome Res. 20, 947–959 (2010).
Stecher, B. et al. Like will to like: abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria. PLoS Pathogens 6, e1000711 (2010).
Bever, J. D., Westover, K. M. & Antonovics, J. Incorporating the soil community into plant population dynamics: the utility of the feedback approach. J. Ecol. 85, 561–573 (1997).
Stark, P. L. & Lee, A. The microbial ecology of the large bowel of breast-fed and formula-fed infants during the 1st year of life. J. Med. Microbiol. 15, 189–203 (1982).
Glover, L. E. & Colgan, S. P. Hypoxia and metabolic factors that influence inflammatory bowel disease pathogenesis. Gastroenterology 140, 1748–1755 (2011).
Goodman, A. L. et al. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc. Natl Acad. Sci. USA 108, 6252–6257 (2011).
Dupont, H. L. Gastrointestinal infections and the development of irritable bowel syndrome. Curr. Opin. Infect. Dis. 24, 503–508 (2011).
Acknowledgements
We would like to thank L. Parfrey, J. Knight and A. Knight for their comments on this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reprints and permissions information is available at www.nature.com/reprints.
Rights and permissions
About this article
Cite this article
Lozupone, C., Stombaugh, J., Gordon, J. et al. Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230 (2012). https://doi.org/10.1038/nature11550
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11550
This article is cited by
-
Exploring new avenues of health protection: plant-derived nanovesicles reshape microbial communities
Journal of Nanobiotechnology (2024)
-
The effect of physical exercise on anticancer immunity
Nature Reviews Immunology (2024)
-
Gut microbiome remodeling and metabolomic profile improves in response to protein pacing with intermittent fasting versus continuous caloric restriction
Nature Communications (2024)
-
Current understanding of the Alzheimer’s disease-associated microbiome and therapeutic strategies
Experimental & Molecular Medicine (2024)
-
Disentangling direct vs indirect effects of microbiome manipulations in a habitat-forming marine holobiont
npj Biofilms and Microbiomes (2024)