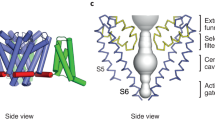
Abstract
Voltage-gated sodium (Nav) channels are essential for the rapid depolarization of nerve and muscle1, and are important drug targets2. Determination of the structures of Nav channels will shed light on ion channel mechanisms and facilitate potential clinical applications. A family of bacterial Nav channels, exemplified by the Na+-selective channel of bacteria (NaChBac)3, provides a useful model system for structure–function analysis. Here we report the crystal structure of NavRh, a NaChBac orthologue from the marine alphaproteobacterium HIMB114 (Rickettsiales sp. HIMB114; denoted Rh), at 3.05 Å resolution. The channel comprises an asymmetric tetramer. The carbonyl oxygen atoms of Thr 178 and Leu 179 constitute an inner site within the selectivity filter where a hydrated Ca2+ resides in the crystal structure. The outer mouth of the Na+ selectivity filter, defined by Ser 181 and Glu 183, is closed, as is the activation gate at the intracellular side of the pore. The voltage sensors adopt a depolarized conformation in which all the gating charges are exposed to the extracellular environment. We propose that NavRh is in an ‘inactivated’ conformation. Comparison of NavRh with NavAb4 reveals considerable conformational rearrangements that may underlie the electromechanical coupling mechanism of voltage-gated channels.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Hille, B. Ion Channels of Excitable Membranes (Sinauer Associates, 2001)
Mantegazza, M., Curia, G., Biagini, G., Ragsdale, D. S. & Avoli, M. Voltage-gated sodium channels as therapeutic targets in epilepsy and other neurological disorders. Lancet Neurol. 9, 413–424 (2010)
Ren, D. et al. A prokaryotic voltage-gated sodium channel. Science 294, 2372–2375 (2001)
Payandeh, J., Scheuer, T., Zheng, N. & Catterall, W. A. The crystal structure of a voltage-gated sodium channel. Nature 475, 353–358 (2011)
Catterall, W. A. The molecular basis of neuronal excitability. Science 223, 653–661 (1984)
Jiang, Y. et al. X-ray structure of a voltage-dependent K+ channel. Nature 423, 33–41 (2003)
Long, S. B., Campbell, E. B. & Mackinnon, R. Voltage sensor of Kv1.2: structural basis of electromechanical coupling. Science 309, 903–908 (2005)
Long, S. B., Tao, X., Campbell, E. B. & MacKinnon, R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature 450, 376–382 (2007)
Butterwick, J. A. & MacKinnon, R. Solution structure and phospholipid interactions of the isolated voltage-sensor domain from KvAP. J. Mol. Biol. 403, 591–606 (2010)
Armstrong, C. M. & Bezanilla, F. Charge movement associated with the opening and closing of the activation gates of the Na channels. J. Gen. Physiol. 63, 533–552 (1974)
Aggarwal, S. K. & MacKinnon, R. Contribution of the S4 segment to gating charge in the Shaker K+ channel. Neuron 16, 1169–1177 (1996)
Seoh, S. A., Sigg, D., Papazian, D. M. & Bezanilla, F. Voltage-sensing residues in the S2 and S4 segments of the Shaker K+ channel. Neuron 16, 1159–1167 (1996)
Vassilev, P. M., Scheuer, T. & Catterall, W. A. Identification of an intracellular peptide segment involved in sodium channel inactivation. Science 241, 1658–1661 (1988)
Armstrong, C. M. & Bezanilla, F. Currents related to movement of the gating particles of the sodium channels. Nature 242, 459–461 (1973)
Hoshi, T., Zagotta, W. N. & Aldrich, R. W. Biophysical and molecular mechanisms of Shaker potassium channel inactivation. Science 250, 533–538 (1990)
Zagotta, W. N., Hoshi, T. & Aldrich, R. W. Restoration of inactivation in mutants of Shaker potassium channels by a peptide derived from ShB. Science 250, 568–571 (1990)
Ulbricht, W. Sodium channel inactivation: molecular determinants and modulation. Physiol. Rev. 85, 1271–1301 (2005)
Todt, H., Dudley, S. C., Jr, Kyle, J. W., French, R. J. & Fozzard, H. A. Ultra-slow inactivation in mu1 Na+ channels is produced by a structural rearrangement of the outer vestibule. Biophys. J. 76, 1335–1345 (1999)
Yellen, G., Sodickson, D., Chen, T. Y. & Jurman, M. E. An engineered cysteine in the external mouth of a K+ channel allows inactivation to be modulated by metal binding. Biophys. J. 66, 1068–1075 (1994)
Durell, S. R. & Guy, H. R. A putative prokaryote voltage-gated Ca2+ channel with only one 6TM motif per subunit. Biochem. Biophys. Res. Commun. 281, 741–746 (2001)
Yue, L., Navarro, B., Ren, D., Ramos, A. & Clapham, D. E. The cation selectivity filter of the bacterial sodium channel, NaChBac. J. Gen. Physiol. 120, 845–853 (2002)
Pavlov, E. et al. The pore, not cytoplasmic domains, underlies inactivation in a prokaryotic sodium channel. Biophys. J. 89, 232–242 (2005)
Harding, M. M. The geometry of metal-ligand interactions relevant to proteins. Acta Crystallogr. D 55, 1432–1443 (1999)
Shannon, R. D. Revised effective ionic radii and systematic studies of interatomic distances in halides and chalcogenides. Acta Crystallogr. A 32, 751–767 (1976)
Hille, B. The hydration of sodium ions crossing the nerve membrane. Proc. Natl Acad. Sci. USA 68, 280–282 (1971)
Armstrong, C. M. & Cota, G. Calcium ion as a cofactor in Na channel gating. Proc. Natl Acad. Sci. USA 88, 6528–6531 (1991)
Schmidt, D., Cross, S. R. & MacKinnon, R. A gating model for the archeal voltage-dependent K+ channel KvAP in DPhPC and POPE:POPG decane lipid bilayers. J. Mol. Biol. 390, 902–912 (2009)
Xiong, W., Li, R. A., Tian, Y. & Tomaselli, G. F. Molecular motions of the outer ring of charge of the sodium channel: do they couple to slow inactivation? J. Gen. Physiol. 122, 323–332 (2003)
Tao, X., Lee, A., Limapichat, W., Dougherty, D. A. & MacKinnon, R. A gating charge transfer center in voltage sensors. Science 328, 67–73 (2010)
DeCaen, P. G., Yarov-Yarovoy, V., Sharp, E. M., Scheuer, T. & Catterall, W. A. Sequential formation of ion pairs during activation of a sodium channel voltage sensor. Proc. Natl Acad. Sci. USA 106, 22498–22503 (2009)
Sawaya, M. R., Pelletier, H., Kumar, A., Wilson, S. H. & Kraut, J. Crystal structure of rat DNA polymerase beta: evidence for a common polymerase mechanism. Science 264, 1930–1935 (1994)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Collaborative Computational Project, 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Schneider, T. R. & Sheldrick, G. M. Substructure solution with SHELXD. Acta Crystallogr. D 58, 1772–1779 (2002)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007)
Cowtan, K. dm: an automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography 31, 34–38 (1994)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 (2002)
DeLano, W. L. The PyMOL Molecular Graphics System. Pymolhttp://www.pymol.org (2002)
Smart, O. S., Goodfellow, J. M. & Wallace, B. A. The pore dimensions of gramicidin A. Biophys. J. 65, 2455–2460 (1993)
Arnold, K., Bordoli, L., Kopp, J. & Schwede, T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 (2006)
Schwede, T., Kopp, J., Guex, N. & Peitsch, M. C. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31, 3381–3385 (2003)
Guex, N. & Peitsch, M. C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18, 2714–2723 (1997)
Echols, N., Milburn, D. & Gerstein, M. MolMovDB: analysis and visualization of conformational change and structural flexibility. Nucleic Acids Res. 31, 478–482 (2003)
Krebs, W. G. & Gerstein, M. The morph server: a standardized system for analyzing and visualizing macromolecular motions in a database framework. Nucleic Acids Res. 28, 1665–1675 (2000)
Brunger, A. T. et al. Crystallography & NMR System: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Brunger, A. T. Version 1.2 of the Crystallography and NMR system. Nature Protocols 2, 2728–2733 (2007)
Acknowledgements
We thank R. MacKinnon at Rockefeller University for discussions and reading the manuscript. We thank L. Feng at Rockefeller University for help. We thank S. Huang and F. Yu at Shanghai Synchrotron Radiation Facility beamline BL17U. K.H. acknowledges SPring-8 beamline BL41XU for proposal 2011A2039. This work was supported by funds from the Ministry of Science and Technology (grant numbers 2009CB918802, 2011CB910501 and 2011CB911102), projects 31125009 and 91017011 of the National Natural Science Foundation of China, and funds from Tsinghua University.
Author information
Authors and Affiliations
Contributions
X.Z., W.R., P.D., X.T., D.E.C. and N.Y. designed experiments. X.Z., W.R., P.D., C.Y., X.T., L.T., J.W., K.H., T.K., J.H., J.W. and N.Y. performed the experiments. X.Z., W.R., P.D., C.Y., X.T., J.W., D.E.C. and N.Y. analysed the data. X.Z., P.D., X.T., C.Y., J.W. and D.E.C. contributed to manuscript preparation. N.Y. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-10, Supplementary Tables 1-2 and the full legend for Supplementary Movie 1. (PDF 3407 kb)
Supplementary Movie
This file contains Supplementary Movie 1 which illustrates the structural basis for gating charge transfer. (MPG 16744 kb)
Rights and permissions
About this article
Cite this article
Zhang, X., Ren, W., DeCaen, P. et al. Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel. Nature 486, 130–134 (2012). https://doi.org/10.1038/nature11054
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11054