Abstract
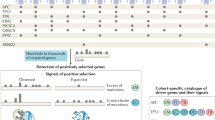
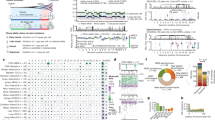
All cancers carry somatic mutations in their genomes. A subset, known as driver mutations, confer clonal selective advantage on cancer cells and are causally implicated in oncogenesis1, and the remainder are passenger mutations. The driver mutations and mutational processes operative in breast cancer have not yet been comprehensively explored. Here we examine the genomes of 100 tumours for somatic copy number changes and mutations in the coding exons of protein-coding genes. The number of somatic mutations varied markedly between individual tumours. We found strong correlations between mutation number, age at which cancer was diagnosed and cancer histological grade, and observed multiple mutational signatures, including one present in about ten per cent of tumours characterized by numerous mutations of cytosine at TpC dinucleotides. Driver mutations were identified in several new cancer genes including AKT2, ARID1B, CASP8, CDKN1B, MAP3K1, MAP3K13, NCOR1, SMARCD1 and TBX3. Among the 100 tumours, we found driver mutations in at least 40 cancer genes and 73 different combinations of mutated cancer genes. The results highlight the substantial genetic diversity underlying this common disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
Data deposits
Genome sequence data have been deposited at the European Genome-phenome Archive under accession number EGAD00001000133. Affymetrix SNP6 data have been deposited under accession number E-MTAB-1110.
Change history
20 June 2012
An initial was added and an affiliation was corrected for author A.L.R.; a hyphen was added to author S.N.-Z.
References
Stratton, M. R., Campbell, P. J. & Futreal, P. A. The cancer genome. Nature 458, 719–724 (2009)
Greenman, C., Wooster, R., Futreal, P. A., Stratton, M. R. & Easton, D. F. Statistical analysis of pathogenicity of somatic mutations in cancer. Genetics 173, 2187–2198 (2006)
Varela, I. et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 469, 539–542 (2011)
Keshet, Y. & Seger, R. The MAP kinase signaling cascades: a system of hundreds of components regulates a diverse array of physiological functions. Methods Mol. Biol. 661, 3–38 (2010)
Su, G. H. et al. Alterations in pancreatic, biliary, and breast carcinomas support MKK4 as a genetically targeted tumor suppressor gene. Cancer Res. 58, 2339–2342 (1998)
Carpten, J. D. et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature 448, 439–444 (2007)
Park, H. S. et al. Akt (protein kinase B) negatively regulates SEK1 by means of protein phosphorylation. J. Biol. Chem. 277, 2573–2578 (2002)
Carnero, A., Blanco-Aparicio, C., Renner, O., Link, W. & Leal, J. F. The PTEN/PI3K/AKT signalling pathway in cancer, therapeutic implications. Curr. Cancer Drug Targets 8, 187–198 (2008)
Wu, G. S. The functional interactions between the MAPK and p53 signaling pathways. Cancer Biol. Ther. 3, 146–151 (2004)
Horlein, A. J. et al. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature 377, 397–404 (1995)
Merrell, K. W. et al. Differential recruitment of nuclear receptor coregulators in ligand-dependent transcriptional repression by estrogen receptor-alpha. Oncogene 30, 1608–1614 (2010)
Jones, S. et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science 330, 228–231 (2010)
Reisman, D., Glaros, S. & Thompson, E. A. The SWI/SNF complex and cancer. Oncogene 28, 1653–1668 (2009)
Wiegand, K. C. et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N. Engl. J. Med. 363, 1532–1543 (2010)
Spirin, K. S. et al. p27/Kip1 mutation found in breast cancer. Cancer Res. 56, 2400–2404 (1996)
Tigli, H., Buyru, N. & Dalay, N. Molecular analysis of the p27/kip1 gene in breast cancer. Mol. Diagn. 9, 17–21 (2005)
Chu, I. M., Hengst, L. & Slingerland, J. M. The Cdk inhibitor p27 in human cancer: prognostic potential and relevance to anticancer therapy. Nature Rev. Cancer 8, 253–267 (2008)
Han, J. et al. Tbx3 improves the germ-line competency of induced pluripotent stem cells. Nature 463, 1096–1100 (2010)
Howard, B. & Ashworth, A. Signalling pathways implicated in early mammary gland morphogenesis and breast cancer. PLoS Genet. 2, e112 (2006)
Bamshad, M. et al. Mutations in human TBX3 alter limb, apocrine and genital development in ulnar-mammary syndrome. Nature Genet. 16, 311–315 (1997)
Fillmore, C. M. et al. Estrogen expands breast cancer stem-like cells through paracrine FGF/Tbx3 signaling. Proc. Natl Acad. Sci. USA 107, 21737–21742 (2010)
Ghoussaini, M. et al. Genome-wide association analysis identifies three new breast cancer susceptibility loci. Nature Genet. 44, 312–318 (2012)
Varghese, J. S. & Easton, D. F. Genome-wide association studies in common cancers–what have we learnt? Curr. Opin. Genet. Dev. 20, 201–209 (2010)
Miller, D. On the nature of susceptibility to cancer. Cancer 46, 1307–1318 (1980)
Schinzel, A. C. & Hahn, W. C. Oncogenic transformation and experimental models of human cancer. Front. Biosci. 13, 71–84 (2008)
Shuck, S. C., Short, E. A. & Turchi, J. J. Eukaryotic nucleotide excision repair: from understanding mechanisms to influencing biology. Cell Res. 18, 64–72 (2008)
Fousteri, M. & Mullenders, L. H. Transcription-coupled nucleotide excision repair in mammalian cells: molecular mechanisms and biological effects. Cell Res. 18, 73–84 (2008)
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010)
Ye, K., Schulz, M. H., Long, Q., Apweiler, R. & Ning, Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 25, 2865–2871 (2009)
Van Loo, P. et al. Allele-specific copy number analysis of tumors. Proc. Natl Acad. Sci. USA 107, 16910–16915 (2010)
Varela, I. et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 469, 539–542 (2011)
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010)
Ye, K., Schulz, M. H., Long, Q., Apweiler, R. & Ning, Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 25, 2865–2871 (2009)
Van Loo, P. et al. Allele-specific copy number analysis of tumors. Proc. Natl Acad. Sci. USA 107, 16910–16915 (2010)
Beroukhim, R. et al. The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905 (2010)
Futreal, P. A. et al. A census of human cancer genes. Nature Rev. Cancer 4, 177–183 (2004)
Greenman, C. et al. Patterns of somatic mutation in human cancer genomes. Nature 446, 153–158 (2007)
Nelder, J. A. &. Wedderburn, R. Generalized linear models. J. R. Stat. Soc. A 135, 370–384 (1972)
Acknowledgements
This work was supported by the Wellcome Trust (grant reference 077012/Z/05/Z) and Breakthrough Breast Cancer. P.J.C. is personally funded through a Wellcome Trust Senior Clinical Research Fellowship (grant reference WT088340MA). P.V.L. is a postdoctoral researcher at the Research Foundation - Flanders (FWO) and is a visiting scientist at the Wellcome Trust Sanger Institute, supported by a travel grant from the FWO. I.V. is supported by a fellowship from The International Human Frontier Science Program Organization. A.-L.B.-D. and A.L. are funded by the Norwegian Research Council, The Norwegian Cancer Society, The Radium Hospital Foundation and Health Region SØ. A.V.S. was supported by an ‘Interface INSERM’ grant. J.S.R.-F. is funded in part by Breakthrough Breast Cancer and is a recipient of the 2010 CRUK Future Leaders Prize. D.E. is a Principal Research Fellow of Cancer Research UK. A.T. receives financial support from the Department of Health via the National Institute for Health Research comprehensive Biomedical Research Centre award to Guy’s and St Thomas’ NHS Foundation Trust in partnership with King’s College London, and from King’s College Hospital NHS Foundation Trust in conjunction with The Experimental Cancer Medicine Centre Initiative jointly funded by Cancer Research UK, the National Institute for Health Research, the Welsh Assembly Government, the HSC R&D Office for Northern Ireland and the Chief Scientist Office, Scotland. C.D. and C.S. received partial funding from the MEDIC foundation and the Fonds National de Recherche Scientifique. J.M. and J.F. are funded in part by a research grant from the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research. The INCa-Synergie facility received support from the Institut National du Cancer, the Fondation Synergie-Lyon-Cancer, the Canceropole Lyon Auverge Rhone Alpes and the Centre Leon Berard. A.C.V. is funded by The Ludwig Institute for Cancer Research. L.v.’t.V. and A. Broeks receive funding from the Dutch Genomics Initiative-Cancer Genomics Center. We also acknowledge support for sample collection, banking and processing from the Biological Resource Center of Institut Curie; the Breakthrough Breast Cancer Unit; P. Watson and the BCCA Tumour Tissue Repository; the Centre for Translational Genomics; A. Lane and P. T. Simpson; the Australian Biospecimens Network; the Breast Unit at Royal Brisbane and Women’s Hospital; the Dana-Farber/Harvard SPORE in breast cancer (reference CA089393); A. M. Sieuwerts; and the Singhealth Tissue Repository, Singapore. We are grateful also for the support of T. B. Tean, and acknowledge the input and guidance of P. Spellman and A. Ashworth.
Author information
Authors and Affiliations
Consortia
Contributions
P.J.S., P.S.T. and H.D. performed analysis of the sequence data, aided by S.N.Z., I.V., G.R.B., L.R.Y., E.P., D.J.M., M.S.-C. and R.R. P.V.L. performed analysis of the SNP6 data. C.G., D.C.W., K.W.L. and D.E. performed the statistical investigations. S. Martin coordinated sample acquisition and pathology review. S. McLaren coordinated sample processing. D.B., A. Butler, J.G., J.H., M.J., A.J., D.J., A.M., K.R. and J.T. performed informatics investigations. A.C., C.L. and L.M performed technical investigations. A.L., OSBREAC, M.T.M.L., C.-Y.S., B.T.K.T., B.W.H., A. Broeks, A.C.V., G. Turashvili, J.M., A.F., P.M., S.-F.C., G. Thomas, S.B., O.M., S.R.L., M.v.d.V., L.v.’t.V., J.F., C.D., C.S., A.T., C.C., J.S.R.-F., S.A.J.R.A., A.V.S., A.-L.B.-D. and A.R. contributed samples, clinical data and scientific advice. P.J.C. and P.A.F. directed the research and contributed to the manuscript. M.R.S. directed the research and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains the Supplementary Statistical Analysis. (PDF 229 kb)
Supplementary Figures
This file contains Supplementary Figures 1-4. (PDF 2173 kb)
Supplementary Tables
This file contains Supplementary Tables 1-7. (XLS 10942 kb)
Rights and permissions
About this article
Cite this article
Stephens, P., Tarpey, P., Davies, H. et al. The landscape of cancer genes and mutational processes in breast cancer. Nature 486, 400–404 (2012). https://doi.org/10.1038/nature11017
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11017
This article is cited by
-
Circulating tumour mutation detection in triple-negative breast cancer as an adjunct to tissue response assessment
npj Breast Cancer (2024)
-
Gene-expression-based T-Cell-to-Stroma Enrichment (TSE) score predicts response to immune checkpoint inhibitors in urothelial cancer
Nature Communications (2024)
-
The impact of PIK3CA mutations and PTEN expression on the effect of neoadjuvant therapy for postmenopausal luminal breast cancer patients
BMC Cancer (2023)
-
Association between vascular ultrasound features and DNA sequencing in breast cancer: a preliminary study
Discover Oncology (2023)
-
Biology of breast cancer brain metastases and novel therapies targeting the blood brain barrier: an updated review
Medical Oncology (2023)