Abstract
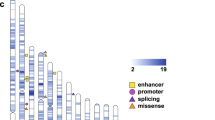
Autism spectrum disorders (ASD) are believed to have genetic and environmental origins, yet in only a modest fraction of individuals can specific causes be identified1,2. To identify further genetic risk factors, here we assess the role of de novo mutations in ASD by sequencing the exomes of ASD cases and their parents (n = 175 trios). Fewer than half of the cases (46.3%) carry a missense or nonsense de novo variant, and the overall rate of mutation is only modestly higher than the expected rate. In contrast, the proteins encoded by genes that harboured de novo missense or nonsense mutations showed a higher degree of connectivity among themselves and to previous ASD genes3 as indexed by protein-protein interaction screens. The small increase in the rate of de novo events, when taken together with the protein interaction results, are consistent with an important but limited role for de novo point mutations in ASD, similar to that documented for de novo copy number variants. Genetic models incorporating these data indicate that most of the observed de novo events are unconnected to ASD; those that do confer risk are distributed across many genes and are incompletely penetrant (that is, not necessarily sufficient for disease). Our results support polygenic models in which spontaneous coding mutations in any of a large number of genes increases risk by 5- to 20-fold. Despite the challenge posed by such models, results from de novo events and a large parallel case–control study provide strong evidence in favour of CHD8 and KATNAL2 as genuine autism risk factors.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Accession codes
Data deposits
Data included in this manuscript have been deposited at dbGaP under accession number phs000298.v1.p1 and is available for download at http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id5phs000298.v1.p1.
References
Lichtenstein, P., Carlstrom, E., Rastam, M., Gillberg, C. & Anckarsater, H. The genetics of autism spectrum disorders and related neuropsychiatric disorders in childhood. Am. J. Psychiatry 167, 1357–1363 (2010)
Hallmayer, J. et al. Genetic heritability and shared environmental factors among twin pairs with autism. Arch. Gen. Psychiatry 68, 1095–1102 (2011)
Betancur, C. Etiological heterogeneity in autism spectrum disorders: more than 100 genetic and genomic disorders and still counting. Brain Res. 1380, 42–77 (2011)
Pinto, D. et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature 466, 368–372 (2010)
Sanders, S. J. et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron 70, 863–885 (2011)
Sebat, J., Levy, D. L. & McCarthy, S. E. Rare structural variants in schizophrenia: one disorder, multiple mutations; one mutation, multiple disorders. Trends Genet. 25, 528–535 (2009)
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010)
DePristo, M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genet. 43, 491–498 (2011)
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010)
Conrad, D. F. et al. Variation in genome-wide mutation rates within and between human families. Nature Genet. 43, 712–714 (2011)
Sanders, S. J. et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature http://dx.doi.org/10.1038/nature10945 (this issue)
Adzhubei, I. A. et al. A method and server for predicting damaging missense mutations. Nature Methods 7, 248–249 (2010)
Kryukov, G. V., Pennacchio, L. A. & Sunyaev, S. R. Most rare missense alleles are deleterious in humans: implications for complex disease and association studies. Am. J. Hum. Genet. 80, 727–739 (2007)
Crow, J. F. The origins, patterns and implications of human spontaneous mutation. Nature Rev. Genet. 1, 40–47 (2000)
Rossin, E. J. et al. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 7, e1001273 (2011)
Lage, K. et al. A large-scale analysis of tissue-specific pathology and gene expression of human disease genes and complexes. Proc. Natl Acad. Sci. USA 105, 20870–20875 (2008)
O’Roak, B. J. et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nature Genet. 43, 585–589 (2011)
O’Roak, B. J. et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature http://dx.doi.org/10.1038/nature10989 (this issue)
Wu, M. C. et al. Rare-variant association testing for sequencing data with the sequence kernel association test. Am. J. Hum. Genet. 89, 82–93 (2011)
Neale, B. M. et al. Testing for an unusual distribution of rare variants. PLoS Genet. 7, e1001322 (2011)
Acknowledgements
This work was directly supported by NIH grants R01MH089208 (M.J.D.), R01 MH089025 (J.D.B.), R01 MH089004 (G.D.S.), R01MH089175 (R.A.G.) and R01 MH089482 (J.S.S.), and supported in part by NIH grants P50 HD055751 (E.H.C.), RO1 MH057881 (B.D.) and R01 MH061009 (J.S.S.). Y.K., G.C. and S.Y. are Seaver Fellows, supported by the Seaver Foundation. We thank T. Lehner, A. Felsenfeld and P. Bender for their support and contribution to the project. We thank S. Sanders and M. State for discussions on the interpretation of de novo events. We thank D. Reich for comments on the abstract and message of the manuscript. We thank E. Lander and D. Altshuler for comments on the manuscript. We acknowledge the assistance of M. Potter, A. McGrew and G. Crockett without whom these studies would not be possible, and Center for Human Genetics Research resources: Computational Genomics Core, Genetic Studies Ascertainment Core and DNA Resources core, supported in part by NIH NCRR grant UL1 RR024975, and the Vanderbilt Kennedy Center for Research on Human Development (P30 HD015052). This work was supported in part by R01MH084676 (S.S.). We acknowledge the clinicians and organizations that contributed to samples used in this study and the particular support of the Mount Sinai School of Medicine, University of Illinois-Chicago, Vanderbilt University, the Autism Genetics Resource Exchange and the institutions of the Boston Autism Consortium. We acknowledge A. Estes and G. Dawson for patient collection/characterization. We acknowledge partial support from U54 HG003273 (R.A.G.) and U54 HG003067 (E. Lander). J.D.B., B.D., M.J.D., R.A.G., A.S., G.D.S. and J.S.S. are lead investigators in the Autism Sequencing Consortium (ASC). The ASC is comprised of groups sharing massively parallel sequencing data in autism. Finally, we are grateful to the many families, without whose participation this project would not have been possible.
Author information
Authors and Affiliations
Contributions
Laboratory work: A.S., C.St., G.C., O.J., Z.P., J.D.B., D.M., I.N., Y.W., L.L., Y.H., S.G., E.L.C., N.G.C. and E.T.G. Data processing: B.M.N., K.E.S., E.L., A.K., J.F., M.F., K.S., T.F., K.G., E.Ba., R.P., M.DeP., S.G., S.Y., V.M., J.L., J.D.B., A.S., C.St., U.N., J.G.R., J.R.W., B.E.B., S.E.L., C.F.L., L.S.W. and O.V. Statistical analysis: B.M.N., L.L., K.E.S., C.Sh., B.F.V., J.M., E.R., S.S., P.P., Y.K., A.M., R.D., C.-F.L., L.-S.W., H.L., T.Z., E.Bo., R.A.G., J.D.B., C.B., E.H.C., J.S.S., G.D.S., B.D., K.R. and M.J.D. Principal Investigators/study design: E.Bo., R.A.G., E.H.C., J.D.B., K.R., B.D., G.D.S., J.S.S. and M.J.D. Y.K., L.L., A.M., K.E.S., A.S. and C.-F.L. contributed equally to this work. E.Bo., J.D.B., E.H.C., B.D., R.A.G., K.R., G.D.S., J.S.S. and M.J.D. are lead investigators of the ARRA Autism Sequencing Collaboration.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Text and Data, Supplementary Figures 1-4, Supplementary Tables 1-10 and additional references. (PDF 1221 kb)
Supplementary Data
This file contains Supplementary Data. (XLS 96 kb)
PowerPoint slides
Rights and permissions
About this article
Cite this article
Neale, B., Kou, Y., Liu, L. et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 485, 242–245 (2012). https://doi.org/10.1038/nature11011
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11011
This article is cited by
-
Statistical methods for assessing the effects of de novo variants on birth defects
Human Genomics (2024)
-
SETD5 haploinsufficiency affects mitochondrial compartment in neural cells
Molecular Autism (2023)
-
Endophenotype trait domains for advancing gene discovery in autism spectrum disorder
Journal of Neurodevelopmental Disorders (2023)
-
Investigation of autism-related transcription factors underlying sex differences in the effects of bisphenol A on transcriptome profiles and synaptogenesis in the offspring hippocampus
Biology of Sex Differences (2023)
-
Early maturation and hyperexcitability is a shared phenotype of cortical neurons derived from different ASD-associated mutations
Translational Psychiatry (2023)