Abstract
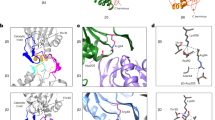
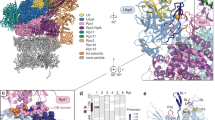
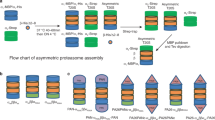
The proteasome is the major ATP-dependent protease in eukaryotic cells, but limited structural information restricts a mechanistic understanding of its activities. The proteasome regulatory particle, consisting of the lid and base subcomplexes, recognizes and processes polyubiquitinated substrates. Here we used electron microscopy and a new heterologous expression system for the lid to delineate the complete subunit architecture of the regulatory particle from yeast. Our studies reveal the spatial arrangement of ubiquitin receptors, deubiquitinating enzymes and the protein unfolding machinery at subnanometre resolution, outlining the substrate’s path to degradation. Unexpectedly, the ATPase subunits within the base unfoldase are arranged in a spiral staircase, providing insight into potential mechanisms for substrate translocation through the central pore. Large conformational rearrangements of the lid upon holoenzyme formation suggest allosteric regulation of deubiquitination. We provide a structural basis for the ability of the proteasome to degrade a diverse set of substrates and thus regulate vital cellular processes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
Data deposits
The cryoelectron microscopy density map for the 26S proteasome can be found at the Electron Microscopy Data Bank under accession number EMD-1992. The negative stain reconstructions of the recombinantly expressed and yeast-purified lid have been assigned accession numbers EMD-1993 and EMD-1994, respectively.
References
Finley, D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78, 477–513 (2009)
Glickman, M. H., Rubin, D. M., Fried, V. A. & Finley, D. The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol. Cell. Biol. 18, 3149–3162 (1998)
Thrower, J. S., Hoffman, L., Rechsteiner, M. & Pickart, C. M. Recognition of the polyubiquitin proteolytic signal. EMBO J. 19, 94–102 (2000)
Groll, M. et al. A gated channel into the proteasome core particle. Nature Struct. Biol. 7, 1062–1067 (2000)
Smith, D. M. et al. Docking of the proteasomal ATPases’ carboxyl termini in the 20S proteasome’s α ring opens the gate for substrate entry. Mol. Cell 27, 731–744 (2007)
Yao, T. & Cohen, R. E. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 419, 403–407 (2002)
Verma, R. et al. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 298, 611–615 (2002)
Tomko, R. J., Jr, Funakoshi, M., Schneider, K., Wang, J. & Hochstrasser, M. Heterohexameric ring arrangement of the eukaryotic proteasomal ATPases: implications for proteasome structure and assembly. Mol. Cell 38, 393–403 (2010)
Rabl, J. et al. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol. Cell 30, 360–368 (2008)
Gillette, T. G., Kumar, B., Thompson, D., Slaughter, C. A. & DeMartino, G. N. Differential roles of the COOH termini of AAA subunits of PA700 (19 S regulator) in asymmetric assembly and activation of the 26 S proteasome. J. Biol. Chem. 283, 31813–31822 (2008)
da Fonseca, P. C. & Morris, E. P. Structure of the human 26S proteasome: subunit radial displacements open the gate into the proteolytic core. J. Biol. Chem. 283, 23305–23314 (2008)
Elsasser, S. et al. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nature Cell Biol. 4, 725–730 (2002)
Gomez, T. A., Kolawa, N., Gee, M., Sweredoski, M. J. & Deshaies, R. J. Identification of a functional docking site in the Rpn1 LRR domain for the UBA-UBL domain protein Ddi1. BMC Biol. 9, 33 (2011)
Leggett, D. S. et al. Multiple associated proteins regulate proteasome structure and function. Mol. Cell 10, 495–507 (2002)
Bohn, S. et al. Structure of the 26S proteasome from Schizosaccharomyces pombe at subnanometer resolution. Proc. Natl Acad. Sci. USA 107, 20992–20997 (2010)
Nickell, S. et al. Insights into the molecular architecture of the 26S proteasome. Proc. Natl Acad. Sci. USA 106, 11943–11947 (2009)
Förster, F. et al. An atomic model AAA-ATPase/20S core particle sub-complex of the 26S proteasome. Biochem. Biophys. Res. Commun. 388, 228–233 (2009)
Xu, P. et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 137, 133–145 (2009)
Inobe, T., Fishbain, S., Prakash, S. & Matouschek, A. Defining the geometry of the two-component proteasome degron. Nature Chem. Biol. 7, 161–167 (2011)
Tian, G. et al. An asymmetric interface between the regulatory and core particles of the proteasome. Nature Struct. Mol. Biol. 18, 1259–1267 (2011)
Effantin, G., Rosenzweig, R., Glickman, M. H. & Steven, A. C. Electron microscopic evidence in support of α-solenoid models of proteasomal subunits Rpn1 and Rpn2. J. Mol. Biol. 386, 1204–1211 (2009)
Zhang, F. et al. Structural insights into the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol. Cell 34, 473–484 (2009)
Hamazaki, J. et al. A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. EMBO J. 25, 4524–4536 (2006)
Schreiner, P. et al. Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature 453, 548–552 (2008)
Glickman, M. H. et al. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell 94, 615–623 (1998)
Verma, R., Oania, R., Graumann, J. & Deshaies, R. J. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell 118, 99–110 (2004)
Glynn, S. E., Martin, A., Nager, A. R., Baker, T. A. & Sauer, R. T. Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine. Cell 139, 744–756 (2009)
Maillard, R. A. et al. ClpX(P) generates mechanical force to unfold and translocate its protein substrates. Cell 145, 459–469 (2011)
Aubin-Tam, M. E., Olivares, A. O., Sauer, R. T., Baker, T. A. & Lang, M. J. Single-molecule protein unfolding and translocation by an ATP-fueled proteolytic machine. Cell 145, 257–267 (2011)
Martin, A., Baker, T. A. & Sauer, R. T. Rebuilt AAA + motors reveal operating principles for ATP-fuelled machines. Nature 437, 1115–1120 (2005)
Hersch, G. L., Burton, R. E., Bolon, D. N., Baker, T. A. & Sauer, R. T. Asymmetric interactions of ATP with the AAA+ ClpX6 unfoldase: allosteric control of a protein machine. Cell 121, 1017–1027 (2005)
Thomsen, N. D. & Berger, J. M. Running in reverse: the structural basis for translocation polarity in hexameric helicases. Cell 139, 523–534 (2009)
Enemark, E. J. & Joshua-Tor, L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature 442, 270–275 (2006)
Riedinger, C. et al. Structure of Rpn10 and its interactions with polyubiquitin chains and the proteasome subunit Rpn12. J. Biol. Chem. 285, 33992–34003 (2010)
Eddins, M. J., Varadan, R., Fushman, D., Pickart, C. M. & Wolberger, C. Crystal structure and solution NMR studies of Lys48-linked tetraubiquitin at neutral pH. J. Mol. Biol. 367, 204–211 (2007)
Cook, W. J., Jeffrey, L. C., Carson, M., Chen, Z. & Pickart, C. M. Structure of a diubiquitin conjugate and a model for interaction with ubiquitin conjugating enzyme (E2). J. Biol. Chem. 267, 16467–16471 (1992)
Bremm, A., Freund, S. M. & Komander, D. Lys11-linked ubiquitin chains adopt compact conformations and are preferentially hydrolyzed by the deubiquitinase Cezanne. Nature Struct. Mol. Biol. 17, 939–947 (2010)
Husnjak, K. et al. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 453, 481–488 (2008)
Hanna, J. et al. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell 127, 99–111 (2006)
Prakash, S., Tian, L., Ratliff, K. S., Lehotzky, R. E. & Matouschek, A. An unstructured initiation site is required for efficient proteasome-mediated degradation. Nature Struct. Mol. Biol. 11, 830–837 (2004)
Verma, R. et al. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol. Biol. Cell 11, 3425–3439 (2000)
Leggett, D. S., Glickman, M. H. & Finley, D. Purification of proteasomes, proteasome subcomplexes, and proteasome-associated proteins from budding yeast. Methods Mol. Biol. 301, 57–70 (2005)
Suloway, C. et al. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151, 41–60 (2005)
Lander, G. C. et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 166, 95–102 (2009)
Tang, G. et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007)
Hohn, M. et al. SPARX, a new environment for Cryo-EM image processing. J. Struct. Biol. 157, 47–55 (2007)
Goddard, T. D., Huang, C. C. & Ferrin, T. E. Visualizing density maps with UCSF Chimera. J. Struct. Biol. 157, 281–287 (2007)
Sone, T., Saeki, Y., Toh-e, A. & Yokosawa, H. Sem1p is a novel subunit of the 26 S proteasome from Saccharomyces cerevisiae. J. Biol. Chem. 279, 28807–28816 (2004)
Kim, H. C. & Huibregtse, J. M. Polyubiquitination by HECT E3s and the determinants of chain type specificity. Mol. Cell. Biol. 29, 3307–3318 (2009)
Mallick, S. P., Carragher, B., Potter, C. S. & Kriegman, D. J. ACE: automated CTF estimation. Ultramicroscopy 104, 8–29 (2005)
Mindell, J. A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003)
Voss, N. R., Yoshioka, C. K., Radermacher, M., Potter, C. S. & Carragher, B. DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J. Struct. Biol. 166, 205–213 (2009)
Sorzano, C. O. et al. XMIPP: a new generation of an open-source image processing package for electron microscopy. J. Struct. Biol. 148, 194–204 (2004)
van Heel, M., Harauz, G., Orlova, E. V., Schmidt, R. & Schatz, M. A new generation of the IMAGIC image processing system. J. Struct. Biol. 116, 17–24 (1996)
Frank, J. et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 (1996)
Heymann, J. B. & Belnap, D. M. Bsoft: image processing and molecular modeling for electron microscopy. J. Struct. Biol. 157, 3–18 (2007)
Acknowledgements
We thank the members of the Martin and Nogales labs for helpful discussions, and G. Cardone for help with local resolution calculations. G.C.L. acknowledges support from Damon Runyon Cancer Research Foundation, M.E.M. acknowledges support by the American Cancer Society grant 121453-PF-11-178-01-TBE, C.B. acknowledges support from the NSF Graduate Research Fellowship. This research was funded in part by the Searle Scholars Program (A.M.), start-up funds from the UC Berkeley MCB Department (A.M.), the NIH grant R01-GM094497-01A1 (A.M.), the Lawrence Berkeley National Laboratory (G.C.L.), and the Howard Hughes Medical Institute (E.N.). Some of the work presented here was conducted at the National Resource for Automated Molecular Microscopy, which is supported by the NIH through the NCRR P41 program (RR017573).
Author information
Authors and Affiliations
Contributions
E.E., M.E.M. and C.B. designed, expressed and purified proteasome constructs, and performed biochemical experiments. G.C.L. performed the electron microscopy, processing and segmentation analysis. All authors contributed to experimental design, data analysis and manuscript preparation.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
The file contains Supplementary Figures 1-12 with legends and Supplementary Table 1. (PDF 9249 kb)
Supplementary Movie 1
The movie shows the 3D reconstruction of the yeast 26S proteasome and the spatial arrangement of all subunits within its regulatory particle, including the ubiquitin receptors and the deubiquitinating enzyme. Docking of crystal structures from homologous proteins reveals a spiral stair case orientation of the six ATPase subunits within the base unfoldase and a horseshoe-shaped arrangement of PCI domains in the lid subcomplex. (MOV 27606 kb)
Rights and permissions
About this article
Cite this article
Lander, G., Estrin, E., Matyskiela, M. et al. Complete subunit architecture of the proteasome regulatory particle. Nature 482, 186–191 (2012). https://doi.org/10.1038/nature10774
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10774