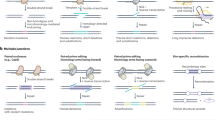
Abstract
Structural variation is widespread in mammalian genomes1,2 and is an important cause of disease3, but just how abundant and important structural variants (SVs) are in shaping phenotypic variation remains unclear4,5. Without knowing how many SVs there are, and how they arise, it is difficult to discover what they do. Combining experimental with automated analyses, we identified 711,920 SVs at 281,243 sites in the genomes of thirteen classical and four wild-derived inbred mouse strains. The majority of SVs are less than 1 kilobase in size and 98% are deletions or insertions. The breakpoints of 160,000 SVs were mapped to base pair resolution, allowing us to infer that insertion of retrotransposons causes more than half of SVs. Yet, despite their prevalence, SVs are less likely than other sequence variants to cause gene expression or quantitative phenotypic variation. We identified 24 SVs that disrupt coding exons, acting as rare variants of large effect on gene function. One-third of the genes so affected have immunological functions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Mills, R. E. et al. Mapping copy number variation by population-scale genome sequencing. Nature 470, 59–65 (2011)
Quinlan, A. R. et al. Genome-wide mapping and assembly of structural variant breakpoints in the mouse genome. Genome Res. 20, 623–635 (2010)
Zhang, F., Gu, W., Hurles, M. E. & Lupski, J. R. Copy number variation in human health, disease, and evolution. Annu. Rev. Genomics Hum. Genet. 10, 451–481 (2009)
Conrad, D. F. et al. Origins and functional impact of copy number variation in the human genome. Nature 464, 704–712 (2010)
Stranger, B. E. et al. Population genomics of human gene expression. Nature Genet. 39, 1217–1224 (2007)
Agam, A. et al. Elusive copy number variation in the mouse genome. PLoS ONE 5, e12839 (2010)
Cahan, P., Li, Y., Izumi, M. & Graubert, T. A. The impact of copy number variation on local gene expression in mouse hematopoietic stem and progenitor cells. Nature Genet. 41, 430–437 (2009)
Henrichsen, C. N. et al. Segmental copy number variation shapes tissue transcriptomes. Nature Genet. 41, 424–429 (2009)
Schadt, E. E. et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nature Genet. 37, 710–717 (2005)
Zhang, F. et al. The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nature Genet. 41, 849–853 (2009)
Ma, J. L., Kim, E. M., Haber, J. E. & Lee, S. E. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol. Cell. Biol. 23, 8820–8828 (2003)
Stankiewicz, P. & Lupski, J. R. Structural variation in the human genome and its role in disease. Annu. Rev. Med. 61, 437–455 (2010)
Stankiewicz, P. & Lupski, J. R. Genome architecture, rearrangements and genomic disorders. Trends Genet. 18, 74–82 (2002)
Hastings, P. J., Ira, G. & Lupski, J. R. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 5, e1000327 (2009)
Huang, G. J. et al. High resolution mapping of expression QTLs in heterogeneous stock mice in multiple tissues. Genome Res. 19, 1133–1140 (2009)
Yalcin, B., Flint, J. & Mott, R. Using progenitor strain information to identify quantitative trait nucleotides in outbred mice. Genetics 171, 673–681 (2005)
Valdar, W. et al. Genome-wide genetic association of complex traits in heterogeneous stock mice. Nature Genet. 38, 879–887 (2006)
Keane, T. M. et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature doi:10.1038/nature10413 (this issue).
Yalcin, B. et al. Commercially available outbred mice for genome-wide association studies. PLoS Genet. 6, e1001085 (2010)
Best, S., Le Tissier, P., Towers, G. & Stoye, J. P. Positional cloning of the mouse retrovirus restriction gene Fv1 . Nature 382, 826–829 (1996)
Boyden, L. M. et al. Skint1, the prototype of a newly identified immunoglobulin superfamily gene cluster, positively selects epidermal γδ T cells. Nature Genet. 40, 656–662 (2008)
Nelson, T. M., Munger, S. D. & Boughter, J. D., Jr Haplotypes at the Tas2r locus on distal chromosome 6 vary with quinine taste sensitivity in inbred mice. BMC Genet. 6, 32 (2005)
Persson, K., Heby, O. & Berger, F. G. The functional intronless S-adenosylmethionine decarboxylase gene of the mouse (Amd-2) is linked to the ornithine decarboxylase gene (Odc) on chromosome 12 and is present in distantly related species of the genus Mus . Mamm. Genome 10, 784–788 (1999)
Wu, B. et al. Mutations in sterol O-acyltransferase 1 (Soat1) result in hair interior defects in AKR/J mice. J. Invest. Dermatol. 130, 2666–2668 (2010)
Tareen, S. U., Sawyer, S. L., Malik, H. S. & Emerman, M. An expanded clade of rodent Trim5 genes. Virology 385, 473–483 (2009)
Taylor, K. et al. Defensin-related peptide 1 (Defr1) is allelic to Defb8 and chemoattracts immature DC and CD4+ T cells independently of CCR6. Eur. J. Immunol. 39, 1353–1360 (2009)
Ye, K., Schulz, M. H., Long, Q., Apweiler, R. & Ning, Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 25, 2865–2871 (2009)
Chen, K. et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nature Methods 6, 677–681 (2009)
Simpson, J. T., McIntyre, R. E., Adams, D. J. & Durbin, R. Copy number variant detection in inbred strains from short read sequence data. Bioinformatics 26, 565–567 (2010)
Manske, H. M. & Kwiatkowski, D. P. LookSeq: a browser-based viewer for deep sequencing data. Genome Res. 19, 2125–2132 (2009)
Acknowledgements
We thank A. Whitley, G. Durrant, A. M. Hammond, D. J. Fabrigar, L. Chen, M. Johannesson, E. Cong and G. Blázquez for helping B.Y. with various laboratory-based work. We also thank C. P. Ponting for comments on the manuscript. This project was supported by The Medical Research Council, UK, and the Wellcome Trust. D.J.A. is supported by Cancer Research UK.
Author information
Authors and Affiliations
Contributions
D.J.A. and J.F. conceived the study and directed the research. J.F. wrote the core of the paper. K.W. and T.K. performed the genome-wide SV discovery and local assembly for SV breakpoint resolution. K.W. carried out the sensitivity and specificity analyses. K.W. and B.Y. liaised regularly to integrate experimental work into genome-wide SV discovery pipeline. This resulted in a highly accurate map of SV across the mouse genome, essential to downstream analyses. A.B., P.H.P., H.W., J.C., R.D. and D.J. carried out experimental work, led by B.Y. A.B. and B.Y. analysed Sanger-based sequencing data, resolved SV breakpoints at nucleotide-level resolution and inferred mechanism of SV formation. M.G. performed the genome-wide SV mechanism of formation and outgroup analysis, with contributions from A.A. and B.Y.; J.F. and A.A. analysed functional impact of SVs on expression and phenotypes. C.N., L.G., J.N., A.A. and R.M. carried out additional analyses. B.Y. characterized function of individual SV examples.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-2 with legends, Supplementary Methods, Supplementary References and Supplementary Tables 1-5. (PDF 722 kb)
PowerPoint slides
Rights and permissions
About this article
Cite this article
Yalcin, B., Wong, K., Agam, A. et al. Sequence-based characterization of structural variation in the mouse genome. Nature 477, 326–329 (2011). https://doi.org/10.1038/nature10432
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10432
This article is cited by
-
An effect of large-scale deletions and duplications on transcript expression
Functional & Integrative Genomics (2023)
-
Interferon signaling promotes tolerance to chromosomal instability during metastatic evolution in renal cancer
Nature Cancer (2023)
-
Murine allele and transgene symbols: ensuring unique, concise, and informative nomenclature
Mammalian Genome (2022)
-
Selection shapes the landscape of functional variation in wild house mice
BMC Biology (2021)
-
Genetics of mouse behavioral and peripheral neural responses to sucrose
Mammalian Genome (2021)