Abstract
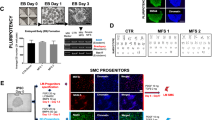
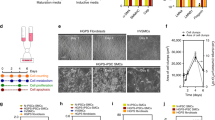
Hutchinson–Gilford progeria syndrome (HGPS) is a rare and fatal human premature ageing disease1,2,3,4,5, characterized by premature arteriosclerosis and degeneration of vascular smooth muscle cells (SMCs)6,7,8. HGPS is caused by a single point mutation in the lamin A (LMNA) gene, resulting in the generation of progerin, a truncated splicing mutant of lamin A. Accumulation of progerin leads to various ageing-associated nuclear defects including disorganization of nuclear lamina and loss of heterochromatin9,10,11,12. Here we report the generation of induced pluripotent stem cells (iPSCs) from fibroblasts obtained from patients with HGPS. HGPS-iPSCs show absence of progerin, and more importantly, lack the nuclear envelope and epigenetic alterations normally associated with premature ageing. Upon differentiation of HGPS-iPSCs, progerin and its ageing-associated phenotypic consequences are restored. Specifically, directed differentiation of HGPS-iPSCs to SMCs leads to the appearance of premature senescence phenotypes associated with vascular ageing. Additionally, our studies identify DNA-dependent protein kinase catalytic subunit (DNAPKcs, also known as PRKDC) as a downstream target of progerin. The absence of nuclear DNAPK holoenzyme correlates with premature as well as physiological ageing. Because progerin also accumulates during physiological ageing6,12,13, our results provide an in vitro iPSC-based model to study the pathogenesis of human premature and physiological vascular ageing.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Worman, H. J., Ostlund, C. & Wang, Y. Diseases of the nuclear envelope. Cold Spring Harb. Perspect. Biol. 2, a000760 (2010)
Burtner, C. R. & Kennedy, B. K. Progeria syndromes and ageing: what is the connection? Nature Rev. Mol. Cell Biol. 11, 567–578 (2010)
Kudlow, B. A., Kennedy, B. K. & Monnat, R. J., Jr Werner and Hutchinson–Gilford progeria syndromes: mechanistic basis of human progeroid diseases. Nature Rev. Mol. Cell Biol. 8, 394–404 (2007)
Merideth, M. A. et al. Phenotype and course of Hutchinson–Gilford progeria syndrome. N. Engl. J. Med. 358, 592–604 (2008)
Davies, B. S., Fong, L. G., Yang, S. H., Coffinier, C. & Young, S. G. The posttranslational processing of prelamin A and disease. Annu. Rev. Genomics Hum. Genet. 10, 153–174 (2009)
Olive, M. et al. Cardiovascular pathology in Hutchinson-Gilford progeria: correlation with the vascular pathology of aging. Arterioscler. Thromb. Vasc. Biol. 10.1161/ATVBAHA.110.209460 (26 August 2010)
Ragnauth, C. D. et al. Prelamin A acts to accelerate smooth muscle cell senescence and is a novel biomarker of human vascular aging. Circulation 121, 2200–2210 (2010)
Varga, R. et al. Progressive vascular smooth muscle cell defects in a mouse model of Hutchinson–Gilford progeria syndrome. Proc. Natl Acad. Sci. USA 103, 3250–3255 (2006)
Pegoraro, G. et al. Ageing-related chromatin defects through loss of the NURD complex. Nature Cell Biol. 11, 1261–1267 (2009)
Scaffidi, P. & Misteli, T. Reversal of the cellular phenotype in the premature aging disease Hutchinson-Gilford progeria syndrome. Nature Med. 11, 440–445 (2005)
Dechat, T. et al. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 22, 832–853 (2008)
Scaffidi, P. & Misteli, T. Lamin A-dependent nuclear defects in human aging. Science 312, 1059–1063 (2006)
McClintock, D. et al. The mutant form of lamin A that causes Hutchinson-Gilford progeria is a biomarker of cellular aging in human skin. PLoS ONE 2, e1269 (2007)
Constantinescu, D., Gray, H. L., Sammak, P. J., Schatten, G. P. & Csoka, A. B. Lamin A/C expression is a marker of mouse and human embryonic stem cell differentiation. Stem Cells 24, 177–185 (2006)
Freberg, C. T., Dahl, J. A., Timoskainen, S. & Collas, P. Epigenetic reprogramming of OCT4 and NANOG regulatory regions by embryonal carcinoma cell extract. Mol. Biol. Cell 18, 1543–1553 (2007)
Peric-Hupkes, D. et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol. Cell 38, 603–613 (2010)
Irizarry, R. A. et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nature Genet. 41, 178–186 (2009)
Huang, W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols 4, 44–57 (2009)
Dennis, G. et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 4, R60 (2003)
McClintock, D., Gordon, L. B. & Djabali, K. Hutchinson–Gilford progeria mutant lamin A primarily targets human vascular cells as detected by an anti-Lamin A G608G antibody. Proc. Natl Acad. Sci. USA 103, 2154–2159 (2006)
Gorenne, I., Kavurma, M., Scott, S. & Bennett, M. Vascular smooth muscle cell senescence in atherosclerosis. Cardiovasc. Res. 72, 9–17 (2006)
Minamino, T. & Komuro, I. Vascular cell senescence: contribution to atherosclerosis. Circ. Res. 100, 15–26 (2007)
Huang, S. et al. Correction of cellular phenotypes of Hutchinson-Gilford progeria cells by RNA interference. Hum. Genet. 118, 444–450 (2005)
Liu, G. H. et al. Regulation of myoblast differentiation by the nuclear envelope protein NET39. Mol. Cell. Biol. 29, 5800–5812 (2009)
Washburn, M. P., Wolters, D. & Yates, J. R., III Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nature Biotechnol. 19, 242–247 (2001)
Ruis, B. L., Fattah, K. R. & Hendrickson, E. A. The catalytic subunit of DNA-dependent protein kinase regulates proliferation, telomere length, and genomic stability in human somatic cells. Mol. Cell. Biol. 28, 6182–6195 (2008)
Li, H., Vogel, H., Holcomb, V. B., Gu, Y. & Hasty, P. Deletion of Ku70, Ku80, or both causes early aging without substantially increased cancer. Mol. Cell. Biol. 27, 8205–8214 (2007)
Espejel, S. et al. Shorter telomeres, accelerated ageing and increased lymphoma in DNA-PKcs-deficient mice. EMBO Rep. 5, 503–509 (2004)
Han, X. et al. Tethering by lamin A stabilizes and targets the ING1 tumour suppressor. Nature Cell Biol. 10, 1333–1340 (2008)
Zhang, J. et al. A human iPSC model of Hutchinson Gilford progeria reveals vascular smooth muscle and mesenchymal stem cell defects. Cell Stem Cell 8, 31–45 (2011)
Candelario, J., Borrego, S., Reddy, S. & Comai, L. Accumulation of distinct prelamin A variants in human diploid fibroblasts differentially affects cell homeostasis. Exp. Cell Res. 317, 319–329 (2011)
Hockemeyer, D. et al. A drug-inducible system for direct reprogramming of human somatic cells to pluripotency. Cell Stem Cell 3, 346–353 (2008)
Vodyanik, M. A., Bork, J. A., Thomson, J. A. & Slukvin, I. I. Human embryonic stem cell-derived CD34+ cells: efficient production in the coculture with OP9 stromal cells and analysis of lymphohematopoietic potential. Blood 105, 617–626 (2005).
Lu, S. J., Ivanova, Y., Feng, Q., Luo, C. & Lanza, R. Hemangioblasts from human embryonic stem cells generate multilayered blood vessels with functional smooth muscle cells. Regen. Med. 4, 37–47 (2009)
Kawamura, T. et al. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 460, 1140–1144 (2009)
Deng, J. et al. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nature Biotechnol. 27, 353–360 (2009)
Bern, M., Goldberg, D., McDonald, W. H. & Yates, J. R., III Automatic quality assessment of peptide tandem mass spectra. Bioinformatics 20 (Suppl 1). i49–i54 (2004)
Eng, J., McCormack, A. & Yates, J. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 (1994)
Peng, J., Elias, J. E., Thoreen, C. C., Licklider, L. J. & Gygi, S. P. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J. Proteome Res. 2, 43–50 (2003)
Sadygov, R. G. et al. Code developments to improve the efficiency of automated MS/MS spectra interpretation. J. Proteome Res. 1, 211–215 (2002)
Debacq-Chainiaux, F., Erusalimsky, J. D., Campisi, J. & Toussaint, O. Protocols to detect senescence-associated beta-galactosidase (SA-βgal) activity, a biomarker of senescent cells in culture and in vivo . Nature Protocols 4, 1798–1806 (2009)
Acknowledgements
We thank L. Comai for providing experimental material, M. Hetzer, J. Karlseder, J.-F. Deleuze, M. J. Barrero, C. Rodriguez Esteban and L. Gerace for helpful discussions, M. Marti for teratoma analysis, M. C. Llach for karyotyping, T. Berggren, M. Lutz, I. Dubova, S. Stewart, R. Dev, M. Li, L. Laricchia-Robbio, A. M. Goebl and J. Kim for technical help, and M. Schwarz for administrative help. G.-H.L. and L.K. were partially supported by a CIRM grant (TG2-01158), J.Q. was partially supported by an AFAR/Ellison Medical Foundation postdoctoral fellowship; A.D.P. was partially supported by a NIH training grant T32 CA009370. This study was supported by grants from NIH R01-DA025779 (K.Z.), and NIH P41 RR011823 (J.Y.); the G. Harold and Leila Y. Mathers Charitable Foundation, Sanofi-Aventis, Ellison Medical Foundation, MICINN and Fundacion Cellex (JCIB).
Author information
Authors and Affiliations
Contributions
G.-H.L. and J.C.I.B. conceived the experiments; G.-H.L., B.Z.B., S.R., D.D., J.Q., S.-L.Y., A.D.P., K.S., L.K., C.W., J.T. and H.L.F. performed the experiments and analysed the data; S.B., I.S.-M., K.Z., J.Y. and J.C.I.B. analysed the data; G.-H.L., S.R., B.Z.B., A.D.P., K.Z. and J.C.I.B. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figures
This file contains Supplementary Figures 1-18 with legends. (PDF 11678 kb)
Supplementary Table 1
This table contains the DNA methylation parameters. (DOC 30 kb)
Supplementary Table 2
This table contains the genes within 10kb of a differentially methylated region (DMR) found between BJ and HGPS fibroblasts. (XLS 356 kb)
Supplementary Table 3
This table contains the genes within 10kb of a differentially methylated region (DMR) found between BJ-iPSCs and HGPS-iPSCs. (XLS 77 kb)
Supplementary Table 4
This table contains the peptides identified by MudPIT for the indicated candidate progerin-associated partners. (DOC 58 kb)
Supplementary Table 5
This table contains the primers used in this study. (DOC 101 kb)
Supplementary Movie 1
In this movie we see the contracting area derived from BJ-iPSC colony following directed differentiation into cardiac tissue. Movies were taken at day 12 of embryoid body development. (MOV 6503 kb)
Supplementary Movie 2
In this movie we see HGPS-iPSCs develop into contractile cardiac tissue with pacemaker activity in vitro. Movies were taken at day 12 of embryoid body development. (MOV 10026 kb)
Rights and permissions
About this article
Cite this article
Liu, GH., Barkho, B., Ruiz, S. et al. Recapitulation of premature ageing with iPSCs from Hutchinson–Gilford progeria syndrome. Nature 472, 221–225 (2011). https://doi.org/10.1038/nature09879
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09879