Abstract
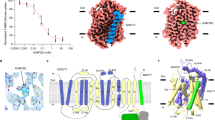
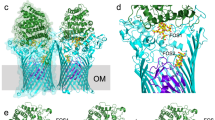
The major facilitator superfamily (MFS) transporters are an ancient and widespread family of secondary active transporters1. In Escherichia coli, the uptake of l-fucose, a source of carbon for microorganisms, is mediated by an MFS proton symporter, FucP2,3. Despite intensive study of the MFS transporters, atomic structure information is only available on three proteins and the outward-open conformation has yet to be captured4,5,6. Here we report the crystal structure of FucP at 3.1 Å resolution, which shows that it contains an outward-open, amphipathic cavity. The similarly folded amino and carboxyl domains of FucP have contrasting surface features along the transport path, with negative electrostatic potential on the N domain and hydrophobic surface on the C domain. FucP only contains two acidic residues along the transport path, Asp 46 and Glu 135, which can undergo cycles of protonation and deprotonation. Their essential role in active transport is supported by both in vivo and in vitro experiments. Structure-based biochemical analyses provide insights into energy coupling, substrate recognition and the transport mechanism of FucP.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Pao, S. S., Paulsen, I. T. & Saier, M. H., Jr Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62, 1–34 (1998)
Bradley, S. A., Tinsley, C. R., Muiry, J. A. & Henderson, P. J. Proton-linked L-fucose transport in Escherichia coli . Biochem. J. 248, 495–500 (1987)
Gunn, F. J., Tate, C. G. & Henderson, P. J. Identification of a novel sugar-H+ symport protein, FucP, for transport of L-fucose into Escherichia coli . Mol. Microbiol. 12, 799–809 (1994)
Abramson, J. et al. Structure and mechanism of the lactose permease of Escherichia coli . Science 301, 610–615 (2003)
Huang, Y., Lemieux, M. J., Song, J., Auer, M. & Wang, D. N. Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli . Science 301, 616–620 (2003)
Yin, Y., He, X., Szewczyk, P., Nguyen, T. & Chang, G. Structure of the multidrug transporter EmrD from Escherichia coli . Science 312, 741–744 (2006)
Becker, D. J. & Lowe, J. B. Fucose: biosynthesis and biological function in mammals. Glycobiology 13, 41R–53R (2003)
Paulsen, I. T., Chauvaux, S., Choi, P. & Saier, M. H., Jr Characterization of glucose-specific catabolite repression-resistant mutants of Bacillus subtilis: identification of a novel hexose:H+ symporter. J. Bacteriol. 180, 498–504 (1998)
Christensen, M. et al. Regulation of expression of the 2-deoxy-D-ribose utilization regulon, deoQKPX, from Salmonella enterica serovar typhimurium. J. Bacteriol. 185, 6042–6050 (2003)
Psakis, G. et al. The sodium-dependent D-glucose transport protein of Helicobacter pylori . Mol. Microbiol. 71, 391–403 (2009)
Horiba, N. et al. Cloning and characterization of a novel Na+-dependent glucose transporter (NaGLT1) in rat kidney. J. Biol. Chem. 278, 14669–14676 (2003)
Law, C. J., Maloney, P. C. & Wang, D. N. Ins and outs of major facilitator superfamily antiporters. Annu. Rev. Microbiol. 62, 289–305 (2008)
Guan, L. & Kaback, H. R. Lessons from lactose permease. Annu. Rev. Biophys. Biomol. Struct. 35, 67–91 (2006)
Smirnova, I., Kasho, V., Sugihara, J. & Kaback, H. R. Probing of the rates of alternating access in LacY with Trp fluorescence. Proc. Natl Acad. Sci. USA 106, 21561–21566 (2009)
Kaback, H. R., Sahin-Toth, M. & Weinglass, A. B. The kamikaze approach to membrane transport. Nature Rev. Mol. Cell Biol. 2, 610–620 (2001)
Kaback, H. R. Structure and mechanism of the lactose permease. C. R. Biol. 328, 557–567 (2005)
Guan, L., Mirza, O., Verner, G., Iwata, S. & Kaback, H. R. Structural determination of wild-type lactose permease. Proc. Natl Acad. Sci. USA 104, 15294–15298 (2007)
Mirza, O., Guan, L., Verner, G., Iwata, S. & Kaback, H. R. Structural evidence for induced fit and a mechanism for sugar/H+ symport in LacY. EMBO J. 25, 1177–1183 (2006)
Gunn, F. J., Tate, C. G., Sansom, C. E. & Henderson, P. J. Topological analyses of the L-fucose-H+ symport protein, FucP, from Escherichia coli . Mol. Microbiol. 15, 771–783 (1995)
Franco, P. J. & Brooker, R. J. Functional roles of Glu-269 and Glu-325 within the lactose permease of Escherichia coli . J. Biol. Chem. 269, 7379–7386 (1994)
Guan, L. & Kaback, H. R. Binding affinity of lactose permease is not altered by the H+ electrochemical gradient. Proc. Natl Acad. Sci. USA 101, 12148–12152 (2004)
Quick, M. et al. Binding of an octylglucoside detergent molecule in the second substrate (S2) site of LeuT establishes an inhibitor-bound conformation. Proc. Natl Acad. Sci. USA 106, 5563–5568 (2009)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Schneider, T. R. & Sheldrick, G. M. Substructure solution with SHELXD. Acta Crystallogr. D 58, 1772–1779 (2002)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007)
Cowtan, K. DM: an automated procedure for phase improvement by density modification. Joint CCP4 ESF-EACBM Newslett. Protein Crystallogr. 31, 34–38 (1994)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 (2002)
DeLano, W. L. PyMOL Molecular Viewer 〈http://www.pymol.org〉 (2002)
Veenhoff, L. M. & Poolman, B. Substrate recognition at the cytoplasmic and extracellular binding site of the lactose transport protein of Streptococcus thermophilus. J. Biol. Chem. 274, 33244–33250 (1999)
Phillips, J. C. et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005)
Brooks, B. R. et al. CHARMM: the biomolecular simulation program. J. Comput. Chem. 30, 1545–1614 (2009)
Acknowledgements
We thank N. Shimizu and T. Kumasaka at Spring-8 beamline BL41XU in Japan, and J. He and S. Huang at the Shanghai Synchrotron Radiation Facility (SSRF) for on-site assistance. This work was supported by funds from the Ministry of Science and Technology (grant number 2009CB918802) and by Tsinghua University 985 Phase II funds. N.Y. acknowledges support from the Yuyuan Foundation and the Li Foundation.
Author information
Authors and Affiliations
Contributions
N.Y. designed the experiments. S.D., L.S., Y.H., F.L. and Y.L. performed the experiments and analysed data. H.G., J.W. and N.Y. analyzed data. N.Y. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
The file contains Supplementary Figures 1-12 with legends, and Supplementary Tables 1-2. (PDF 2230 kb)
Rights and permissions
About this article
Cite this article
Dang, S., Sun, L., Huang, Y. et al. Structure of a fucose transporter in an outward-open conformation. Nature 467, 734–738 (2010). https://doi.org/10.1038/nature09406
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09406
This article is cited by
-
Cultivation of marine bacteria of the SAR202 clade
Nature Communications (2023)
-
Identification of a major facilitator superfamily protein that is beneficial to L-lactic acid production by Bacillus coagulans at low pH
BMC Microbiology (2022)
-
X-ray crystallography reveals molecular recognition mechanism for sugar binding in a melibiose transporter MelB
Communications Biology (2021)
-
Cloning and Molecular Characterization of CcNRT2.1/CcNAR2, a Putative Inducible High Affinity Nitrate Transport System in Capsicum chinense Jacq. Roots
Tropical Plant Biology (2020)
-
Mechanistic basis of L-lactate transport in the SLC16 solute carrier family
Nature Communications (2019)