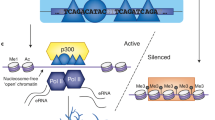
Abstract
We used genome-wide sequencing methods to study stimulus-dependent enhancer function in mouse cortical neurons. We identified ∼12,000 neuronal activity-regulated enhancers that are bound by the general transcriptional co-activator CBP in an activity-dependent manner. A function of CBP at enhancers may be to recruit RNA polymerase II (RNAPII), as we also observed activity-regulated RNAPII binding to thousands of enhancers. Notably, RNAPII at enhancers transcribes bi-directionally a novel class of enhancer RNAs (eRNAs) within enhancer domains defined by the presence of histone H3 monomethylated at lysine 4. The level of eRNA expression at neuronal enhancers positively correlates with the level of messenger RNA synthesis at nearby genes, suggesting that eRNA synthesis occurs specifically at enhancers that are actively engaged in promoting mRNA synthesis. These findings reveal that a widespread mechanism of enhancer activation involves RNAPII binding and eRNA synthesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
Primary accessions
Gene Expression Omnibus
Data deposits
Sequencing data have been submitted to the GEO repository under accession numbers GSE21161 (for all ChIP-Seq and RNA-Seq data) and HM047267 (for circularized Arc enhancer RNA). The bigWig files for genome browser visualization are posted online (see Supplementary Table 6).
References
Banerji, J., Rusconi, S. & Schaffner, W. Expression of a β-globin gene is enhanced by remote SV40 DNA sequences. Cell 27, 299–308 (1981)
Greer, P. L. & Greenberg, M. E. From synapse to nucleus: calcium-dependent gene transcription in the control of synapse development and function. Neuron 59, 846–860 (2008)
Heintzman, N. D. et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nature Genet. 39, 311–318 (2007)
Visel, A. et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457, 854–858 (2009)
Xi, H. et al. Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome. PLoS Genet. 3, e136 (2007)
Park, P. J. ChIP-seq: advantages and challenges of a maturing technology. Nature Rev. Genet. 10, 669–680 (2009)
Barski, A. et al. High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 (2007)
Robertson, A. G. et al. Genome-wide relationship between histone H3 lysine 4 mono- and tri-methylation and transcription factor binding. Genome Res. 18, 1906–1917 (2008)
Chowdhury, S. et al. Arc/Arg3.1 interacts with the endocytic machinery to regulate AMPA receptor trafficking. Neuron 52, 445–459 (2006)
Plath, N. et al. Arc/Arg3.1 is essential for the consolidation of synaptic plasticity and memories. Neuron 52, 437–444 (2006)
Shepherd, J. D. et al. Arc/Arg3.1 mediates homeostatic synaptic scaling of AMPA receptors. Neuron 52, 475–484 (2006)
Waltereit, R. et al. Arg3.1/Arc mRNA induction by Ca2+ and cAMP requires protein kinase A and mitogen-activated protein kinase/extracellular regulated kinase activation. J. Neurosci. 21, 5484–5493 (2001)
Kawashima, T. et al. Synaptic activity-responsive element in the Arc/Arg3.1 promoter essential for synapse-to-nucleus signaling in activated neurons. Proc. Natl Acad. Sci. USA 106, 316–321 (2009)
Pintchovski, S. A., Peebles, C. L., Kim, H. J., Verdin, E. & Finkbeiner, S. The serum response factor and a putative novel transcription factor regulate expression of the immediate-early gene Arc/Arg3.1 in neurons. J. Neurosci. 29, 1525–1537 (2009)
Flavell, S. W. & Greenberg, M. E. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu. Rev. Neurosci. 31, 563–590 (2008)
Lin, Y. et al. Activity-dependent regulation of inhibitory synapse development by Npas4. Nature 455, 1198–1204 (2008)
Kee, B. L., Arias, J. & Montminy, M. R. Adaptor-mediated recruitment of RNA polymerase II to a signal-dependent activator. J. Biol. Chem. 271, 2373–2375 (1996)
Brodsky, A. S. et al. Genomic mapping of RNA polymerase II reveals sites of co-transcriptional regulation in human cells. Genome Biol. 6, R64 (2005)
Kim, T. H. et al. A high-resolution map of active promoters in the human genome. Nature 436, 876–880 (2005)
Koch, F., Jourquin, F., Ferrier, P. & Andrau, J. C. Genome-wide RNA polymerase II: not genes only!. Trends Biochem. Sci. 33, 265–273 (2008)
Szutorisz, H., Dillon, N. & Tora, L. The role of enhancers as centres for general transcription factor recruitment. Trends Biochem. Sci. 30, 593–599 (2005)
Abeel, T., Saeys, Y., Rouze, P. & Van de Peer, Y. ProSOM: core promoter prediction based on unsupervised clustering of DNA physical profiles. Bioinformatics 24, i24–i31 (2008)
Ling, J. et al. HS2 enhancer function is blocked by a transcriptional terminator inserted between the enhancer and the promoter. J. Biol. Chem. 279, 51704–51713 (2004)
Zhao, H. & Dean, A. An insulator blocks spreading of histone acetylation and interferes with RNA polymerase II transfer between an enhancer and gene. Nucleic Acids Res. 32, 4903–4919 (2004)
Heintzman, N. D. et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459, 108–112 (2009)
Gerber, M. & Shilatifard, A. Transcriptional elongation by RNA polymerase II and histone methylation. J. Biol. Chem. 278, 26303–26306 (2003)
Kawashima, T. et al. Synaptic activity-responsive element in the Arc/Arg3.1 promoter essential for synapse-to-nucleus signaling in activated neurons. Proc. Natl Acad. Sci. USA 106, 316–321 (2009)
Couttlet, P. et al. Messenger RNA deadenylation precedes decapping in mammalian cells. Proc. Natl Acad. Sci. USA 94, 5628–5633 (1997)
Flavell, S. W. et al. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron 60, 1022–1038 (2008)
Kharchenko, P. V., Tolstorukov, M. Y. & Park, P. J. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nature Biotechnol. 26, 1351–1359 (2008)
Rozowsky, J. et al. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nature Biotechnol. 27, 66–75 (2009)
Robertson, A. G. et al. Genome-wide relationship between histone H3 lysine 4 mono- and tri-methylation and transcription factor binding. Genome Res. 18, 1906–1917 (2008)
Skellam, J. G. The frequency distribution of the difference between two Poisson variates belonging to different populations. J. R. Stat. Soc. A 109, 296 (1946)
Efron, B. Microarrays, empirical Bayes and the two-groups model. Stat. Sci. 23, 1–22 (2008)
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Statist. Soc. Ser. B. Methodol. 57, 289–300 (1995)
Jothi, R., Cuddapah, S., Barski, A., Cui, K. & Zhao, K. Genome-wide identification of in vivo protein–DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 36, 5221–5231 (2008)
Ooe, N. et al. Identification of a novel basic helix-loop-helix-PAS factor, NXF, reveals a Sim2 competitive, positive regulatory role in dendritic-cytoskeleton modulator drebrin gene expression. Mol. Cell. Biol. 24, 608–616 (2004)
Lin, Y. et al. Activity-dependent regulation of inhibitory synapse development by Npas4. Nature 455, 1198–1204 (2008)
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protocols 4, 44–57 (2009)
Dennis, G. et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4, 3 (2003)
Acknowledgements
We thank members of the Greenberg laboratory for discussions and for critical reading of the manuscript. We thank S. Vasquez for preparing dissociated mouse cortical neurons. We thank L. Hu for generating antibodies. We thank the Molecular Genetics Core Facility at Children’s Hospital Boston, including H. Schneider and S. Burgess, for operation of their SOLiD 3.0 sequencer (I.D.D.R.C). We thank the support and R&D teams at Life Technologies including S. Ranade, R. David, J. Ni, C. Barbacioru, M. Barker, G. Costa and K. McKernan. M.E.G. acknowledges the support of the Nancy Lurie Marks Family Foundation. We thank M. Dehoff for technical support in the Arc knockout experiments. This work was supported by the National Institutes of Health grants NS028829 (M.E.G.), R21EY019710 (G.K.), DP2OD006461 (G.K.) and MH-053608 (P.F.W.). This work was also supported by The Lefler postdoctoral fellowship (T-K.K.) and The Jane Coffin Childs Memorial Funds (T-K.K.), The Helen Hay Whitney postdoctoral fellowship (J.M.G.), The Children’s Hospital Ophthalmology Foundation (G.K.), The Whitehall Foundation (G.K.), and The Klingenstein Fund (G.K.)
Author information
Authors and Affiliations
Contributions
Author Contributions T-K.K., J.M.G. and M.E.G. conceived and designed experiments. T-K.K., J.M.G., M.H., G.K. and M.E.G. wrote the manuscript. T-K.K. optimized the protocol for ChIP-Seq library preparation to be suitable for the SOLiD sequencer and made all ChIP-Seq libraries used in this study. S.K. invented the library construction methodology used for all RNA sequencing reported here. J.M.G., A.M.C. and E.M.-P. made all RNA-Seq libraries. M.H., J.M.G. and D.A.H. performed bioinformatic analyses. K.B.-H. carried out the SOLiD bead preparation and sequencing. T-K.K., J.W., P.F.W. and A.M.C. performed the Arc knockout experiment. D.M.B. performed the luciferase experiments. M.L. performed the RNA circularization experiment. H.B. provided the pArc7000 plasmid. D.K. provided the Arc knockout mouse. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figures
DESCRIPTION (PDF 5982 kb)
This file contains Supplementary Figures 1-11 with legends.
This zipped file comprises Supplementary Tables as follows: Supplementary Table S1 shows the number of CBP sites that were removed at each step of filtering in order to produce a list of high-confidence enhancers. Supplementary Table S2 shows ChIP-Seq, RNA-Seq, and other data associated with each CBP peak, with ~41,000 CBP peaks as rows and nearly 200 columns of information about each peak. Supplementary Tables S3a and S3b show lists of TF peaks found in the 2hour KCl stimulated and unstimulated conditions, with positive values for any given factor indicating the presence of a peak. Supplementary Table S4 shows the number of extragenic enhancers that have TFs, RNAPII, or eRNAs detected, as well as the number of enhancers with any two of these features detected. Supplementary Table S5 shows the number of ChIP-Seq/RNA-Seq reads for each experiment and the antibody used for each ChIP-Seq experiment. Supplementary Table S6 contains text that can be pasted into the UCSC Genome Browser to display the raw ChIP-Seq/RNA-Seq sequencing data using the mm9 mouse genome. Supplementary Table S7 shows DAVID analysis of the genes whose promoters bind CREB. Supplementary Table S8 shows a list of genes with expression changes that were significant following 6 hours KCl stimulation, based on RNA-Seq biological replicate 1. Supplementary Tables S9a and 9b show DAVID analysis of strongly up-and down-regulated genes. Supplementary Table S10 shows primers used for RT-qPCR validation. (ZIP 14681 kb)
Rights and permissions
About this article
Cite this article
Kim, TK., Hemberg, M., Gray, J. et al. Widespread transcription at neuronal activity-regulated enhancers. Nature 465, 182–187 (2010). https://doi.org/10.1038/nature09033
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09033
This article is cited by
-
Transcription directionality is licensed by Integrator at active human promoters
Nature Structural & Molecular Biology (2024)
-
MYC activity at enhancers drives prognostic transcriptional programs through an epigenetic switch
Nature Genetics (2024)
-
Identification and characterization of transcribed enhancers during cerebellar development through enhancer RNA analysis
BMC Genomics (2023)
-
Downregulated NPAS4 in multiple brain regions is associated with major depressive disorder
Scientific Reports (2023)
-
JUN-induced super-enhancer RNA forms R-loop to promote nasopharyngeal carcinoma metastasis
Cell Death & Disease (2023)