Abstract
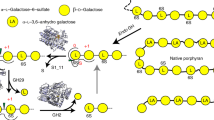
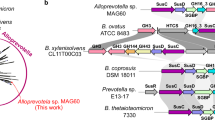
Gut microbes supply the human body with energy from dietary polysaccharides through carbohydrate active enzymes, or CAZymes1, which are absent in the human genome. These enzymes target polysaccharides from terrestrial plants that dominated diet throughout human evolution2. The array of CAZymes in gut microbes is highly diverse, exemplified by the human gut symbiont Bacteroides thetaiotaomicron3, which contains 261 glycoside hydrolases and polysaccharide lyases, as well as 208 homologues of susC and susD-genes coding for two outer membrane proteins involved in starch utilization1,4. A fundamental question that, to our knowledge, has yet to be addressed is how this diversity evolved by acquiring new genes from microbes living outside the gut. Here we characterize the first porphyranases from a member of the marine Bacteroidetes, Zobellia galactanivorans, active on the sulphated polysaccharide porphyran from marine red algae of the genus Porphyra. Furthermore, we show that genes coding for these porphyranases, agarases and associated proteins have been transferred to the gut bacterium Bacteroides plebeius isolated from Japanese individuals5. Our comparative gut metagenome analyses show that porphyranases and agarases are frequent in the Japanese population6 and that they are absent in metagenome data7 from North American individuals. Seaweeds make an important contribution to the daily diet in Japan (14.2 g per person per day)8, and Porphyra spp. (nori) is the most important nutritional seaweed, traditionally used to prepare sushi9,10. This indicates that seaweeds with associated marine bacteria may have been the route by which these novel CAZymes were acquired in human gut bacteria, and that contact with non-sterile food may be a general factor in CAZyme diversity in human gut microbes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Cantarel, B. L. et al. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D238 (2009)
Ley, R. E., Lozupone, C. A., Hamady, M., Knight, R. & Gordon, J. I. Worlds within worlds: evolution of the vertebrate gut microbiota. Nature Rev. Microbiol. 6, 776–778 (2008)
Xu, J. et al. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299, 2074–2076 (2003)
Martens, E. C., Koropatkin, N. M., Smith, T. J. & Gordon J. I Complex glycan catabolism by the human gut microbiota: the Bacteroidetes Sus-like paradigm. J. Biol. Chem. 284, 24673–24677 (2009)
Kitahara, M., Sakamoto, M., Ike, M., Sakata, S. & Benno, Y. Bacteroides plebeius sp. nov. and Bacteroides coprocola sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 55, 2143–2147 (2005)
Kurokawa, K. et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 14, 169–181 (2007)
Turnbaugh, P. J. et al. A core gut microbiome in obese and lean twins. Nature 457, 480–484 (2009)
Fukuda, S. et al. Pattern of dietary fiber intake among the Japanese general population. Eur. J. Clin. Nutr. 61, 99–103 (2007)
Nisizawa, K., Noda, H., Kikuchi, R. & Watanabe, T. The main seaweed foods in Japan. Hydrobiologia 151–152, 5–29 (1987)
Mc Hugh, D. J. in FAO Fisheries Technical Paper No 441 (FAO, 2003)
Kloareg, B. & Quatrano, R. S. Structure of the cell walls of marine algae and ecophysiological functions of the matrix polysaccharides. Oceanogr. Mar. Biol Annu. Rev. 26, 259–315 (1988)
Michel, G., Nyvall-Collen, P., Barbeyron, T., Czjzek, M. & Helbert, W. Bioconversion of red seaweed galactans: a focus on bacterial agarases and carrageenases. Appl. Microbiol. Biotechnol. 71, 23–33 (2006)
Gilbert, H. J., Stalbrand, H. & Brumer, H. How the walls come crumbling down: recent structural biochemistry of plant polysaccharide degradation. Curr. Opin. Plant Biol. 11, 338–348 (2008)
Barbeyron, T. et al. Zobellia galactanovorans gen. nov., sp. nov., a marine species of Flavobacteriaceae isolated from a red alga, and classification of [Cytophaga] uliginosa (ZoBell and Upham 1944) Reichenbach 1989 as Zobellia uliginosa gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 51, 985–987 (2001)
Barbeyron, T., Gerard, A., Potin, P., Henrissat, B. & Kloareg, B. The κ-carrageenase of the marine bacterium Cytophaga drobachiensis. Structural and phylogenetic relationships within family-16 glycoside hydrolases. Mol. Biol. Evol. 15, 528–537 (1998)
Jam, M. et al. The endo-β-agarases AgaA and AgaB from the marine bacterium Zobellia galactanivorans: two paralogue enzymes with different molecular organizations and catalytic behaviours. Biochem. J. 385, 703–713 (2005)
Allouch, J. et al. The three-dimensional structures of two β-agarases. J. Biol. Chem. 278, 47171–47180 (2003)
Maciel, J. S. et al. Structural characterization of cold extracted fraction of soluble sulfated polysaccharide from the red seaweed Gracilaria birdiae . Carbohydr. Polym. 71, 559–565 (2008)
Turvey, J. R. & Rees, D. A. Isolation of l-galactose-6-sulphate from a seaweed polysaccharide. Nature 189, 831–832 (1961)
Anderson, N. S. & Rees, D. A. Porphyran — a polysaccharide with a masked repeating structure. J. Chem. Soc. 5880–5887 (1965)
Garcillán-Barcia, M. P., Francia, M. V. & de la Cruz, F. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol. Rev. 33, 657–687 (2009)
Zhong, Z. et al. Sequence analysis of a 101-kilobase plasmid required for agar degradation by a Microscilla isolate. Appl. Environ. Microbiol. 67, 5771–5779 (2001)
Fisher, R. A. On the interpretation of χ2 from contingency tables, and the calculation of P. J. R. Stat. Soc. 85, 87–94 (1922)
Kuo, C. H. & Ochman, H. Inferring clocks when lacking rocks: the variable rates of molecular evolution in bacteria. Biol. Direct 4, 35 (2009)
Ishihara, K., Oyamada, C., Matsushima, R., Murata, M. & Muraoka, T. Inhibitory effect of porphyran, prepared from dried “nori”, on contact hypersensitivity in mice. Biosci. Biotechnol. Biochem. 69, 1824–1830 (2005)
Kidby, D. K. & Davidson, D. J. A convenient ferricyanide estimation of reducing sugars in the nanomole range. Anal. Biochem. 55, 321–325 (1973)
Pape, T. & Schneider, T. R. HKL2MAP: a graphical user interface for macromolecular phasing with SHELX programs. J. Appl. Cryst. 37, 843–844 (2004)
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997)
Potterton, L. et al. Developments in the CCP4 molecular-graphics project. Acta Crystallogr. D 60, 2288–2294 (2004)
Boraston, A. B., Bolam, D. N., Gilbert, H. J. & Davies, G. J. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382, 769–781 (2004)
Studier, F. W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 (2005)
Kabsch, W. Evaluation of single-crystal X-ray-diffraction data from a position-sensitive detector. J. Appl. Cryst. 21, 916–924 (1988)
Schneider, T. R. & Sheldrick, G. M. Substructure solution with SHELXD. Acta Crystallogr. D 58, 1772–1779 (2002)
Potterton, E., Briggs, P., Turkenburg, M. & Dodson, E. A graphical user interface to the CCP4 program suite. Acta Crystallogr. D 59, 1131–1137 (2003)
Leslie, A. G. W. & Powell, H. R. in Evolving Methods for Macromolecular Crystallography Vol. 245 41–51 (Springer, 2007)
Starr, C. M., Masada, R. I., Hague, C., Skop, E. & Klock, J. C. Fluorophore-assisted carbohydrate electrophoresis in the separation, analysis, and sequencing of carbohydrates. J. Chromatogr. A 720, 295–321 (1996)
Katoh, K., Misawa, K., Kuma, K.-i. & Miyata, T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 (2002)
Gouet, P., Robert, X. & Courcelle, E. ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 31, 3320–3323 (2003)
Michel, G. et al. The κ-carrageenase of P. carrageenovora features a tunnel-shaped active site: a novel insight in the evolution of Clan-B glycoside hydrolases. Structure 9, 513–525 (2001)
Guindon, S. & Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704 (2003)
Kumar, S., Tamura, K. & Nei, M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5, 150–163 (2004)
Acknowledgements
We thank B. Kloareg and C. de Vargas for critical discussions and reading of the manuscript and M. Jam, A. Jeudy and D. Freudenreich for technical assistance. The ‘Marine Plants and Biomolecules’ laboratory is funded by the French national research centre (Centre National de la Recherche Scientifique) and the University Marie Curie; J.-H.H. was supported by a European Marie Curie PhD grant; this work was also funded by the ‘Region Bretagne’ through the program Marine 3D. G.M. was supported by the GIS ‘Genomique Marine’ and the French Research Ministry (ACI Young Researcher). We thank the beamline scientists and staff at the European Synchrotron Radiation Facilities for technical support during data collections, the NMR Service, University Bretagne Occidentale, for access to the Bruker NMR spectrometer and Genoscope for sequencing the Z. galactanivorans genome.
Author Contributions J.-H.H. cloned, purified and crystallized the enzymes and extracted polysaccharides; J.-H.H. and M.C. collected data and solved the crystal structures; G.C. and J.-H.H. purified and characterized oligosaccharides; G.C. and W.H. performed the NMR analysis; G.M., T.B. and J.-H.H. performed the bioinformatic analysis; M.C., T.B., G.M. and J.-H.H. designed the study; J.-H.H., M.C. and G.M. analysed the data and wrote the paper. All authors discussed the results and commented on the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Supplementary information
Supplementary Information
This file contains Supplementary Notes A - C with References, Supplementary Tables 1-7 and Supplementary Figures 1-18 with Legends. (PDF 7309 kb)
Rights and permissions
About this article
Cite this article
Hehemann, JH., Correc, G., Barbeyron, T. et al. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature 464, 908–912 (2010). https://doi.org/10.1038/nature08937
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature08937