Abstract
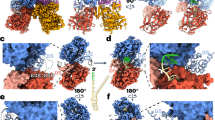
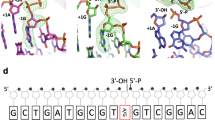
Escherichia coli AlkB and its human homologues ABH2 and ABH3 repair DNA/RNA base lesions by using a direct oxidative dealkylation mechanism. ABH2 has the primary role of guarding mammalian genomes against 1-meA damage by repairing this lesion in double-stranded DNA (dsDNA), whereas AlkB and ABH3 preferentially repair single-stranded DNA (ssDNA) lesions and can repair damaged bases in RNA. Here we show the first crystal structures of AlkB–dsDNA and ABH2–dsDNA complexes, stabilized by a chemical cross-linking strategy. This study reveals that AlkB uses an unprecedented base-flipping mechanism to access the damaged base: it squeezes together the two bases flanking the flipped-out one to maintain the base stack, explaining the preference of AlkB for repairing ssDNA lesions over dsDNA ones. In addition, the first crystal structure of ABH2, presented here, provides a structural basis for designing inhibitors of this human DNA repair protein.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
Primary accessions
Protein Data Bank
Data deposits
Atomic coordinates are deposited in Protein Data Bank under accession numbers 3BKZ ((Mn/2KG) AlkB–DNA1), 3BI3 ((Mn/2KG) AlkB–DNA5), 3BIE ((Mn/2KG)AlkB–DNA4), 3BTX (ABH2–DNA2), 3BTZ (ABH2–DNA3), 3BU0 ((Mn/2KG)ABH2–DNA2), 3BTY (ABH2–DNA6) and 3BUC ((Mn/2KG)ABH2–DNA6).
References
Friedberg, E. C., Walker, G. C. & Siede, W. DNA Repair and Mutagenesis Ch. 1 (ASM Press, Washington DC, 1995)
Wood, R. D., Mitchell, M., Sgouros, J. & Lindahl, T. Human DNA repair genes. Science 291, 1284–1289 (2001)
Sedgwick, B. Repairing DNA-methylation damage. Nature Rev. Mol. Cell Biol. 5, 148–157 (2004)
Mishina, Y. & He, C. Oxidative dealkylation DNA repair mediated by the monomuclear non-heme iron AlkB proteins. J. Inorg. Biochem. 100, 670–678 (2006)
Lindahl, T., Sedgewick, B., Sekiguchi, M. & Nakabeppu, Y. Regulation and expression of the adaptive response to alkylating agents. Annu. Rev. Biochem. 57, 133–157 (1988)
Falnes, P. O., Johansen, R. F. & Seeberg, E. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli . Nature 419, 178–182 (2002)
Trewick, S. C., Henshaw, T. F., Hausinger, R. P., Lindahl, T. & Sedgwick, B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature 419, 174–178 (2002)
Aravind, L. & Koonin, E. V. The DNA-repair protein AlkB, EGL-9, and leprecan define new families of 2-oxoglutarate- and iron-dependent dioxygenases. Genome Biol. 2, research0007 (2001)
Koivisto, P., Robins, P., Lindahl, T. & Sedgwick, B. Demethylation of 3-methylthymine in DNA by bacterial and human DNA dioxygenases. J. Biol. Chem. 279, 40470–40474 (2004)
Falnes, P. O. Repair of 3-methylthymine and 1-methylguanine lesions by bacterial and human AlkB proteins. Nucleic Acids Res. 32, 6260–6267 (2004)
Delaney, J. C. & Essigmann, J. M. Mutagenesis, genotoxicity, and repair of 1-methyladenine, 3-alkylcytosines, 1-methylguanine, and 3-methylthymine in alkB Escherichia coli . Proc. Natl Acad. Sci. USA 101, 14051–14056 (2004)
Mishina, Y., Yang, C.-G. & He, C. Direct repair of the exocyclic DNA adduct 1,N6-ethenoadenine by the DNA repair AlkB proteins. J. Am. Chem. Soc. 127, 14594–14595 (2005)
Delaney, J. C. et al. AlkB reverses etheno DNA lesions caused by lipid oxidation in vitro and in vivo . Nature Struct. Mol. Biol. 12, 855–860 (2005)
Duncan, T. et al. Reversal of DNA alkylation damage by two human dioxygenases. Proc. Natl Acad. Sci. USA 99, 16660–16665 (2002)
Aas, P. A. et al. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature 421, 859–863 (2003)
Lee, D.-H. et al. Repair of methylation damage in DNA and RNA by mammalian AlkB homologues. J. Biol. Chem. 280, 39448–39459 (2005)
Gerken, T. et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science 318, 1469–1472 (2007)
Ringvoll, J. et al. Repair deficient mice reveal mABH2 as the primary oxidative demethylase for repairing 1meA and 3meC lesions in DNA. EMBO J. 25, 2189–2198 (2006)
Yu, B. et al. Crystal structures of catalytic complexes of the oxidative DNA/RNA repair enzyme AlkB. Nature 439, 879–884 (2006)
Sundheim, O. et al. Human ABH3 structure and key residues for oxidative demethylation to reverse DNA/RNA damage. EMBO J. 25, 3389–3397 (2006)
Mishina, Y., Chen, L. X. & He, C. Preparation and characterization of the native iron(II)-containing DNA repair AlkB protein directly from Escherichia coli . J. Am. Chem. Soc. 126, 16930–16936 (2004)
Dinglay, S., Trewick, S. C., Lindahl, T. & Sedgwick, B. Defective processing of methylated single-stranded DNA by E. coli AlkB mutants. Genes Dev. 14, 2097–2105 (2000)
Huang, H., Chopra, R., Verdine, G. L. & Harrison, S. C. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase at 2.7 Å resolution: implications of comformational changes for polymerization and inhibition mechanism. Science 282, 1669–1675 (1998)
Verdine, G. L. & Norman, D. P. G. Covalent trapping of protein–DNA complexes. Annu. Rev. Biochem. 72, 337–366 (2003)
Mishina, Y. & He, C. Probing the structure and function of the Escherichia coli DNA alkylation repair AlkB protein through chemical cross-linking. J. Am. Chem. Soc. 125, 8730–8731 (2003)
Duguid, E. M., Mishina, Y. & He, C. How do DNA repair proteins locate potential base lesions? A chemical crosslinking method to investigate the damage-searching mechanism of O6-alkylguanine-DNA alkyltransferases. Chem. Biol. 10, 827–835 (2003)
Mishina, Y., Lee, C. H. & He, C. Interaction of human and bacterial AlkB proteins with DNA as probed through chemical cross-linking studies. Nucleic Acids Res. 32, 1548–1554 (2004)
Schofield, C. J. & Zhang, Z. Structural and mechanistic studies on 2-oxoglutarate-dependent oxygenases and related enzymes. Curr. Opin. Struct. Biol. 9, 722–731 (1999)
Lange, S. J. & Que, L. Oxygen activating nonheme iron enzymes. Curr. Opin. Chem. Biol. 2, 159–172 (1998)
Elkins, J. M. et al. X-ray crystal structure of Escherichia coli taurine/α-ketoglutarate dioxygenase complexed to ferrous iron and substrates. Biochemistry 41, 5185–5192 (2002)
Lau, A. Y., Scharer, O. D., Samson, L., Verdine, G. L. & Ellenberger, T. Crystal structure of a human alkylbase-DNA repair enzyme complexed to DNA: mechanismf for nucleotide flipping and base excision. Cell 95, 249–258 (1998)
Fromme, J. C., Banerjee, A., Huang, S. J. & Verdine, G. L. Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase. Nature 427, 652–656 (2004)
Banerjee, A., Santos, W. L. & Verdine, G. L. Structure of a DNA glycocylase searching for lesions. Science 311, 1153–1157 (2006)
Slupphaug, G. et al. A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature 384, 87–92 (1996)
Bruner, S. D., Norman, D. P. G. & Verdine, G. L. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature 403, 859–866 (2000)
Parker, J. B. et al. Enzymatic capture of an extrahelical thymine in the search for uracil in DNA. Nature 449, 433–437 (2007)
Kissinger, C. R., Gehlhaar, D. K. & Fogel, D. B. Rapid automated molecular replacement by evolutionary search. Acta Crystallogr. D 55, 484–491 (1999)
Read, R. J. Pushing the boundaries of molecular replacement with maximum likelihood. Erratum. Acta Crystallogr. D 59, 404 (2003)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D. 50, 760–763 (1994)
Otwinowski, Z. W. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Terwilliger, T. C. & Berendzen, J. Automated MAD and MIR structure solution. Acta Crystallogr. D 55, 849–861 (1999)
Brünger, A. T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
DeLano, W. L. The PyMOL molecular graphics system (DeLano Scientific, Palo Alto, California, 2002)
Acknowledgements
We thank: R. Zhang and other beamline staff for assistance with data collection; Y. Luo and the Proteomics and Informatics Services Facility at Research Resources Center (University of Illinois at Chicago) for liquid chromatography–mass spectrometry analysis; and X. Yang, H. Chen and P. R. Chen for discussions. We also thank T. Lindahl and B. Sedgwick for the gift of the abh2 gene. This work was supported by National Institutes of Health (GM071440 to C.H. and a PCBio fellowship for C.T.S.), the W. M. Keck Foundation Distinguished Young Scholar in Medical Research Program (C.H.), and the Arnold and Mabel Beckman Foundation Young Investigator Program (C.H.). Data collection was performed at beamlines 19BM (Structure Biology Center) and 14BM (BioCARS) at the Advanced Photon Source at Argonne National Laboratory; financial support for these beamlines comes from the National Institutes of Health and the United States Department of Energy.
Author Contributions C.-G.Y. and C.Y. solved all AlkB–dsDNA and ABH2–dsDNA structures with help from E.M.D. (crystallography), C.T.S. (initial construct of ABH2 and crystallography) and X.J. (biochemistry). P.A.R. contributed to protein crystallography. C.H. designed the overall project and wrote the manuscript with C.-G.Y. and C.Y. All authors discussed results and commented on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary information
This file contains Supplementary Tables 1-2, Supplementary Figures 1-16 with Legends, Supplementary Method and Result, and additional references (PDF 7316 kb)
Rights and permissions
About this article
Cite this article
Yang, CG., Yi, C., Duguid, E. et al. Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA. Nature 452, 961–965 (2008). https://doi.org/10.1038/nature06889
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature06889