Abstract
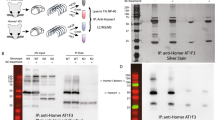
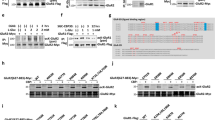
The small ubiquitin-like modifier protein (SUMO) regulates transcriptional activity and the translocation of proteins across the nuclear membrane1. The identification of SUMO substrates outside the nucleus is progressing2 but little is yet known about the wider cellular role of protein SUMOylation. Here we report that in rat hippocampal neurons multiple SUMOylation targets are present at synapses and we show that the kainate receptor subunit GluR6 is a SUMO substrate. SUMOylation of GluR6 regulates endocytosis of the kainate receptor and modifies synaptic transmission. GluR6 exhibits low levels of SUMOylation under resting conditions and is rapidly SUMOylated in response to a kainate but not an N-methyl-D-aspartate (NMDA) treatment. Reducing GluR6 SUMOylation using the SUMO-specific isopeptidase SENP-1 prevents kainate-evoked endocytosis of the kainate receptor. Furthermore, a mutated non-SUMOylatable form of GluR6 is not endocytosed in response to kainate in COS-7 cells. Consistent with this, electrophysiological recordings in hippocampal slices demonstrate that kainate-receptor-mediated excitatory postsynaptic currents are decreased by SUMOylation and enhanced by deSUMOylation. These data reveal a previously unsuspected role for SUMO in the regulation of synaptic function.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Seeler, J. S. & Dejean, A. Nuclear and unclear functions of SUMO. Nature Rev. Mol. Cell Biol. 4, 690– 699 (2003)
Wilson, V. G. & Rosas-Acosta, G. Wrestling with SUMO in a new arena. Sci. STKE 2005, pe32 (2005)
Lerma, J. Roles and rules of kainate receptors in synaptic transmission. Nature Rev. Neurosci. 4, 481– 495 (2003)
Jaskolski, F., Coussen, F. & Mulle, C. Subcellular localization and trafficking of kainate receptors. Trends Pharmacol. Sci. 26, 20– 26 (2005)
Hilgarth, R. S. et al. Regulation and function of SUMO modification. J. Biol. Chem. 279, 53899– 53902 (2004)
Johnson, E. S. Protein modification by SUMO. Annu. Rev. Biochem. 73, 355– 382 (2004)
Suzuki, T. et al. A new 30-kDa ubiquitin-related SUMO-1 hydrolase from bovine brain. J. Biol. Chem. 274, 31131– 31134 (1999)
Hay, R. T. SUMO: a history of modification. Mol. Cell 18, 1– 12 (2005)
Martin, S. & Henley, J. M. Activity-dependent endocytic sorting of kainate receptors to recycling or degradation pathways. EMBO J. 23, 4749– 4759 (2004)
Gong, L., Millas, S., Maul, G. G. & Yeh, E. T. Differential regulation of sentrinized proteins by a novel sentrin-specific protease. J. Biol. Chem. 275, 3355– 3359 (2000)
Rajan, S., Plant, L. D., Rabin, M. L., Butler, M. H. & Goldstein, S. A. Sumoylation silences the plasma membrane leak K+ channel K2P1. Cell 121, 37– 47 (2005)
Uchimura, Y., Nakao, M. & Saitoh, H. Generation of SUMO-1 modified proteins in E. coli: towards understanding the biochemistry/structural biology of the SUMO-1 pathway. FEBS Lett. 564, 85– 90 (2004)
Vignes, M. & Collingridge, G. L. The synaptic activation of kainate receptors. Nature 388, 179– 182 (1997)
Castillo, P. E., Malenka, R. C. & Nicoll, R. A. Kainate receptors mediate a slow postsynaptic current in hippocampal CA3 neurons. Nature 388, 182– 188 (1997)
Mulle, C. et al. Altered synaptic physiology and reduced susceptibility to kainate- induced seizures in GluR6-deficient mice. Nature 392, 601– 605 (1998)
Luscher, C. et al. Role of AMPA receptor cycling in synaptic transmission and plasticity. Neuron 24, 649– 658 (1999)
Borgdorff, A. J. & Choquet, D. Regulation of AMPA receptor lateral movements. Nature 417, 649– 653 (2002)
Comerford, K. M. et al. Small ubiquitin-related modifier-1 modification mediates resolution of CREB-dependent responses to hypoxia. Proc. Natl Acad. Sci. USA 100, 986– 991 (2003)
Zhang, J. et al. c-fos regulates neuronal excitability and survival. Nature Genet. 30, 416– 420 (2002)
Hirbec, H. et al. Rapid and differential regulation of AMPA and kainate receptors at hippocampal mossy fibre synapses by PICK1 and GRIP. Neuron 37, 625– 638 (2003)
Coussen, F. et al. Co-assembly of two GluR6 kainate receptor splice variants within a functional protein complex. Neuron 47, 555– 566 (2005)
Nishimune, A. et al. NSF binding to GluR2 regulates synaptic transmission. Neuron 21, 87– 97 (1998)
Kim, J. et al. Sindbis vector SINrep(nsP2S726): a tool for rapid heterologous expression with attenuated cytotoxicity in neurons. J. Neurosci. Methods 133, 81– 90 (2004)
Hirbec, H., Martin, S. & Henley, J. M. Syntenin is involved in the developmental regulation of neuronal membrane architecture. Mol. Cell. Neurosci. 28, 737– 746 (2005)
Fleck, M. W., Cornell, E. & Mah, S. J. Amino-acid residues involved in glutamate receptor 6 kainate receptor gating and desensitization. J. Neurosci. 23, 1219– 1227 (2003)
Mah, S. J., Cornell, E., Mitchell, N. A. & Fleck, M. W. Glutamate receptor trafficking: endoplasmic reticulum quality control involves ligand binding and receptor function. J. Neurosci. 25, 2215– 2225 (2005)
Lee, H. K., Kameyama, K., Huganir, R. L. & Bear, M. F. NMDA induces long-term synaptic depression and dephosphorylation of the GluR1 subunit of AMPA receptors in hippocampus. Neuron 21, 1151– 1162 (1998)
Daw, M. I. et al. PDZ proteins interacting with C-terminal GluR2/3 are involved in a PKC- dependent regulation of AMPA receptors at hippocampal synapses. Neuron 28, 873– 886 (2000)
Acknowledgements
We acknowledge the Wellcome Trust (J.M.H.), the MRC (J.R.M. and J.M.H.) and the EU (J.M.H. from a GRIPPANT contract) for financial support. We thank G. Hodgkinson for validation electrophysiology experiments on recombinant GluR6 in HEK cells, and S. Correa for some preliminary immuno cytochemistry experiments. We also thank M. Fleck for anti-GluR6 antibody, S. Goldstein for the SENP-1 constructs, C. Mulle for the pcDNA3-myc-GluR6 and H. Saitoh for the bacterial SUMOylation system. We are grateful to M. Ashby, G. Banting, Z. Bashir, T. Bouschet, G. Collingridge, J. Hanley, J. Isaac, F. Jaskolski, A. Randall and D. Stephens for commenting on the manuscript.
Author Contributions S.M. and A. N. are co-first authors; J.R.M. and J.M.H. are co-last authors. S.M. performed surface expression, biochemistry and cell imaging assays in cell culture. A.N. made the original observation that GluR6a is a SUMOylation substrate, performed biochemistry on brain extracts and performed all molecular biological experiments. J.R.M. performed the electrophysiology experiments and co-wrote the manuscript. J.M.H. provided team leadership, project management and wrote the manuscript. All authors contributed to hypothesis development, experimental design and data interpretation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Supplementary information
Supplementary Figures
This file contains Supplementary Figures 1-11 with Legends. (PDF 3951 kb)
Rights and permissions
About this article
Cite this article
Martin, S., Nishimune, A., Mellor, J. et al. SUMOylation regulates kainate-receptor-mediated synaptic transmission. Nature 447, 321–325 (2007). https://doi.org/10.1038/nature05736
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature05736
This article is cited by
-
Knocking Down PIAS3 Reduces H2O2-induced Oxidative Stress Injury in HT22 Cells
Cell Biochemistry and Biophysics (2024)
-
SUMOylation of PDGF receptor α affects signaling via PLCγ and STAT3, and cell proliferation
BMC Molecular and Cell Biology (2023)
-
Bidirectional regulation of synaptic SUMOylation by Group 1 metabotropic glutamate receptors
Cellular and Molecular Life Sciences (2022)
-
Impaired dopamine metabolism in Parkinson’s disease pathogenesis
Molecular Neurodegeneration (2019)
-
Insulin-dependent GLUT4 trafficking is not regulated by protein SUMOylation in L6 myocytes
Scientific Reports (2019)