Abstract
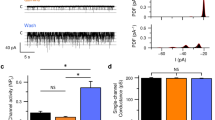
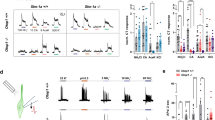
Mammals taste many compounds yet use a sensory palette consisting of only five basic taste modalities: sweet, bitter, sour, salty and umami (the taste of monosodium glutamate)1,2. Although this repertoire may seem modest, it provides animals with critical information about the nature and quality of food. Sour taste detection functions as an important sensory input to warn against the ingestion of acidic (for example, spoiled or unripe) food sources1,2,3. We have used a combination of bioinformatics, genetic and functional studies to identify PKD2L1, a polycystic-kidney-disease-like ion channel4, as a candidate mammalian sour taste sensor. In the tongue, PKD2L1 is expressed in a subset of taste receptor cells distinct from those responsible for sweet, bitter and umami taste. To examine the role of PKD2L1-expressing taste cells in vivo, we engineered mice with targeted genetic ablations of selected populations of taste receptor cells. Animals lacking PKD2L1-expressing cells are completely devoid of taste responses to sour stimuli. Notably, responses to all other tastants remained unaffected, proving that the segregation of taste qualities even extends to ionic stimuli. Our results now establish independent cellular substrates for four of the five basic taste modalities, and support a comprehensive labelled-line mode of taste coding at the periphery5,6,7,8,9,10. Notably, PKD2L1 is also expressed in specific neurons surrounding the central canal of the spinal cord. Here we demonstrate that these PKD2L1-expressing neurons send projections to the central canal, and selectively trigger action potentials in response to decreases in extracellular pH. We propose that these cells correspond to the long-sought components of the cerebrospinal fluid chemosensory system11. Taken together, our results suggest a common basis for acid sensing in disparate physiological settings.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Lindemann, B. Receptors and transduction in taste. Nature 413, 219–225 (2001)
Kinnamon, S. C. & Margolskee, R. F. Mechanisms of taste transduction. Curr. Opin. Neurobiol. 6, 506–513 (1996)
DeSimone, J. A., Lyall, V., Heck, G. L. & Feldman, G. M. Acid detection by taste receptor cells. Respir. Physiol. 129, 231–245 (2001)
Wu, G. et al. Identification of PKD2L, a human PKD2-related gene: tissue-specific expression and mapping to chromosome 10q25. Genomics 54, 564–568 (1998)
Adler, E. et al. A novel family of mammalian taste receptors. Cell 100, 693–702 (2000)
Nelson, G. et al. Mammalian sweet taste receptors. Cell 106, 381–390 (2001)
Nelson, G. et al. An amino-acid taste receptor. Nature 416, 199–202 (2002)
Zhang, Y. et al. Coding of sweet, bitter, and umami tastes: different receptor cells sharing similar signaling pathways. Cell 112, 293–301 (2003)
Zhao, G. Q. et al. The receptors for mammalian sweet and umami taste. Cell 115, 255–266 (2003)
Mueller, K. L. et al. The receptors and coding logic for bitter taste. Nature 434, 225–229 (2005)
Vigh, B. et al. The system of cerebrospinal fluid-contacting neurons. Its supposed role in the nonsynaptic signal transmission of the brain. Histol. Histopathol. 19, 607–628 (2004)
Lyall, V. et al. The mammalian amiloride-insensitive non-specific salt taste receptor is a vanilloid receptor-1 variant. J. Physiol. (Lond.) 558, 147–159 (2004)
Vinnikova, A. K. et al. Na+-H+ exchange activity in taste receptor cells. J. Neurophysiol. 91, 1297–1313 (2004)
Matsunami, H., Montmayeur, J. P. & Buck, L. B. A family of candidate taste receptors in human and mouse. Nature 404, 601–604 (2000)
Chandrashekar, J. et al. T2Rs function as bitter taste receptors. Cell 100, 703–711 (2000)
Li, X. et al. Human receptors for sweet and umami taste. Proc. Natl Acad. Sci. USA 99, 4692–4696 (2002)
Nauli, S. M. et al. Polycystins 1 and 2 mediate mechanosensation in the primary cilium of kidney cells. Nature Genet. 33, 129–137 (2003)
Delmas, P. Polycystins: polymodal receptor/ion-channel cellular sensors. Pflugers Arch. 451, 264–276 (2005)
Clapham, D. E. TRP channels as cellular sensors. Nature 426, 517–524 (2003)
Murakami, M. et al. Genomic organization and functional analysis of murine PKD2L1. J. Biol. Chem. 280, 5626–5635 (2005)
LopezJimenez, N. D. et al. Two members of the TRPP family of ion channels, Pkd1l3 and Pkd2l1, are co-expressed in a subset of taste receptor cells. J. Neurochem. 98, 68–77 (2006)
Hoon, M. A. et al. Putative mammalian taste receptors: a class of taste-specific GPCRs with distinct topographic selectivity. Cell 96, 541–551 (1999)
Collier, R. J. Diphtheria toxin: mode of action and structure. Bacteriol. Rev. 39, 54–85 (1975)
Brockschnieder, D. et al. Cell depletion due to diphtheria toxin fragment A after Cre-mediated recombination. Mol. Cell. Biol. 24, 7636–7642 (2004)
Perez, C. A. et al. A transient receptor potential channel expressed in taste receptor cells. Nature Neurosci. 5, 1169–1176 (2002)
Lahiri, S. & Forster, R. E. II CO2/H+ sensing: peripheral and central chemoreception. Int. J. Biochem. Cell Biol. 35, 1413–1435 (2003)
Richerson, G. B., Wang, W., Hodges, M. R., Dohle, C. I. & Diez-Sampedro, A. Homing in on the specific phenotype(s) of central respiratory chemoreceptors. Exp. Physiol. 90, 259–266, 266–269 (2005)
Gosgnach, S. et al. V1 spinal neurons regulate the speed of vertebrate locomotor outputs. Nature 440, 215–219 (2006)
Lee, E. C. et al. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73, 56–65 (2001)
Novak, A., Guo, C., Yang, W., Nagy, A. & Lobe, C. G. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis 28, 147–155 (2000)
Acknowledgements
We thank L. Feng for help with expression studies, A. Becker for generation of antibodies, D. Cowan for sequencing, and K. Briedis for bioinformatics. We especially thank Y. Zhang for introducing us to the spinal cord slice preparation, and superb technical guidance and help with equipment and animals. We thank members of the Zuker laboratory for valuable comments. This research was supported in part by a grant from the National Institute on Deafness and Other Communication Disorders to C.S.Z. and the intramural research program of the NIH, NIDCR (N.J.P.R.). X.C. is a fellow of the HFS program and D.T. is supported by an Emmy-Noether grant of the Deutsche Forschungsgemeinschaft. C.S.Z. is an investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Supplementary information
Supplementary Figures
This file contains Supplementary Figures 1–3 (PDF 149 kb)
Rights and permissions
About this article
Cite this article
Huang, A., Chen, X., Hoon, M. et al. The cells and logic for mammalian sour taste detection. Nature 442, 934–938 (2006). https://doi.org/10.1038/nature05084
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature05084
This article is cited by
-
Alkaline taste sensation through the alkaliphile chloride channel in Drosophila
Nature Metabolism (2023)
-
Cerebrospinal fluid-contacting neurons: multimodal cells with diverse roles in the CNS
Nature Reviews Neuroscience (2023)
-
Polycystic kidney disease 2-like 1 channel contributes to the bitter aftertaste perception of quinine
Scientific Reports (2023)
-
The proton channel OTOP1 is a sensor for the taste of ammonium chloride
Nature Communications (2023)
-
Ascl1-expressing cell differentiation in initially developed taste buds and taste organoids
Cell and Tissue Research (2023)