Abstract
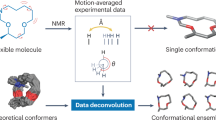
We present a protocol for the experimental determination of ensembles of protein conformations that represent simultaneously the native structure and its associated dynamics. The procedure combines the strengths of nuclear magnetic resonance spectroscopy—for obtaining experimental information at the atomic level about the structural and dynamical features of proteins—with the ability of molecular dynamics simulations to explore a wide range of protein conformations. We illustrate the method for human ubiquitin in solution and find that there is considerable conformational heterogeneity throughout the protein structure. The interior atoms of the protein are tightly packed in each individual conformation that contributes to the ensemble but their overall behaviour can be described as having a significant degree of liquid-like character. The protocol is completely general and should lead to significant advances in our ability to understand and utilize the structures of native proteins.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Frauenfelder, H., Sligar, S. G. & Wolynes, P. G. The energy landscapes and motions on proteins. Science 254, 1598–1603 (1991)
Rasmussen, B. F., Stock, A. M., Ringe, D. & Petsko, G. A. Crystalline ribonuclease A loses function below the dynamical transition at 220 K. Nature 357, 423–424 (1992)
Karplus, M. & McCammon, J. A. Molecular dynamics simulations of biomolecules. Nature Struct. Biol. 9, 646–652 (2002)
Eisenmesser, E. Z., Bosco, D. A., Akke, M. & Kern, D. Enzyme dynamics during catalysis. Science 295, 1520–1523 (2002)
Wong, C. F. & McCammon, J. A. Protein flexibility and computer-aided drug design. Annu. Rev. Pharmacol. Toxicol. 43, 31–45 (2003)
Benkovic, S. J. & Hammes-Schiffer, S. A perspective on enzyme catalysis. Science 301, 1196–1202 (2003)
Kay, L. E. Protein dynamics from NMR. Nature Struct. Biol. 5, 513–517 (1998)
Best, R. B. & Vendruscolo, M. Determination of ensembles of structures consistent with NMR order parameters. J. Am. Chem. Soc. 126, 8090–8091 (2004)
Tjandra, N., Feller, S. E., Pastor, R. W. & Bax, A. Rotational diffusion anisotropy of human ubiquitin from 15N NMR relaxation. J. Am. Chem. Soc. 117, 12562–12566 (1995)
Tjandra, N. & Bax, A. Direct measurement of distances and angles in biomolecules by NMR in a dilute liquid crystalline medium. Science 278, 1111–1114 (1997)
Cornilescu, G., Marquardt, J. L., Ottiger, M. & Bax, A. Validation of protein structure from anisotropic carbonyl chemical shifts in a dilute liquid crystalline phase. J. Am. Chem. Soc. 120, 6836–6837 (1998)
Ottiger, M. & Bax, A. How tetrahedral are methyl groups in proteins? A liquid crystal NMR study. J. Am. Chem. Soc. 121, 4690–4695 (1999)
Lee, A. L., Flynn, P. F. & Wand, A. J. Comparison of 2H and 13C NMR relaxation techniques for the study of protein methyl group dynamics in solution. J. Am. Chem. Soc. 121, 2891–2902 (1999)
Peti, W., Meiler, J., Brüschweiler, R. & Griesinger, C. Model-free analysis of protein backbone motion from residual dipolar couplings. J. Am. Chem. Soc. 124, 5822–5833 (2002)
Chou, J. J., Case, D. A. & Bax, A. Insights into the mobility of methyl-bearing side chains in proteins from 3J CC and 3J CN couplings. J. Am. Chem. Soc. 125, 8959–8966 (2003)
Clore, G. M. & Schwieters, C. D. How much backbone motion in ubiquitin is required to account for dipolar coupling data measured in multiple alignment media as assessed by independent cross-validation? J. Am. Chem. Soc. 126, 2923–2938 (2004)
Kitao, A. & Wagner, G. A space-time structure determination of human CD2 reveals the CD58-binding mode. Proc. Natl Acad. Sci. USA 97, 2064–2068 (2000)
Scheek, R. M., Torda, A. E., Kemmink, J. & van Gunsteren, W. F. in Computational Aspects of the Study of Biological Macromolecules by Nuclear Magnetic Resonance Spectroscopy (eds Hoch, J. C., Redfield, C. & Poulsen, F. M.) 209–217 (Plenum, New York, 1991)
Bonvin, A. M. J. J., Rullmann, J. A. C., Lamerichs, R. M. J. N., Boelens, R. & Kaptein, R. ‘Ensemble’ iterative relaxation matrix approach: A new NMR refinement protocol applied to the solution structure of crambin. Proteins 15, 385–400 (1993)
Choy, W. Y. & Forman-Kay, J. D. Calculation of ensembles of structures representing the unfolded state of an SH3 domain. J. Mol. Biol. 308, 1011–1032 (2001)
Vijay-Kumar, S., Bugg, C. E. & Cook, W. J. Structure of ubiquitin refined at 1.8 Å resolution. J. Mol. Biol. 194, 531–544 (1987)
Mierke, D. F., Scheek, R. M. & Kessler, H. Coupling constants as restraints in ensemble driven dynamics. Biopolymers 34, 559–563 (1994)
Stillinger, F. H. & Stillinger, D. K. Computational study of transition dynamics in 55-atom clusters. J. Chem. Phys. 93, 6013–6024 (1990)
Zhou, Y., Vitkup, D. & Karplus, M. Native proteins are surface-molten solids: Application of the Lindemann criterion for the solid versus liquid state. J. Mol. Biol. 285, 1371–1375 (1999)
DePristo, M. A., de Bakker, P. I. & Blundell, T. L. Heterogeneity and inaccuracy in protein structures solved by X-ray crystallography. Structure 12, 831–838 (2004)
Hoch, J. C., Dobson, C. M. & Karplus, M. Vicinal coupling constants and protein dynamics. Biochemistry 24, 3831–3841 (1984)
Best, R. B., Clarke, J. & Karplus, M. The origin of protein sidechain order parameter distributions. J. Am. Chem. Soc. 126, 7734–7735 (2004)
Markley, J. L. et al. Recommendations for the presentation of NMR structures of proteins and nucleic acids. J. Mol. Biol. 280, 933–952 (1998)
Richards, F. M. The interpretation of protein structures: Total volume, group volume distribution and packing density. J. Mol. Biol. 82, 1–14 (1974)
Pontius, J., Richelle, J. & Wodak, S. J. Deviation from standard atomic volumes as a quality measure for protein crystal structure. J. Mol. Biol. 264, 121–136 (1996)
Guerois, R., Nielsen, J. E. & Serrano, L. Predicting changes in the stability of proteins and protein complexes: A study of more than 1000 mutations. J. Mol. Biol. 320, 369–387 (2002)
Buck, M. & Karplus, M. Internal and overall peptide group motion in proteins: Molecular dynamics simulations for lysozyme compared with results from X-ray and NMR spectroscopy. J. Am. Chem. Soc. 121, 9645–9658 (1999)
Vendruscolo, M., Paci, E., Dobson, C. M. & Karplus, M. Rare fluctuations of native proteins sampled by equilibrium hydrogen exchange. J. Am. Chem. Soc. 125, 15686–15687 (2003)
Lee, A. L. & Wand, A. J. Microscopic origins of entropy, heat capacity and the glass transition in proteins. Nature 411, 501–504 (2001)
Yang, D. & Kay, L. E. Contributions to conformational entropy arising from bond vector fluctuations measured from NMR-derived order parameters: Application to protein folding. J. Mol. Biol. 263, 369–382 (1996)
Johnson, E. C., Lazar, G. A., Desjarlais, J. R. & Handel, T. M. Solution structure and dynamics of a designed hydrophobic core variant of ubiquitin. Struct. Fold. Des. 7, 967–976 (1999)
Benítez-Cardoza, C. G. et al. Exploring sequence/folding space: Folding studies on multiple hydrophobic core mutants of ubiquitin. Biochemistry 43, 5195–5203 (2004)
Lindorff-Larsen, K. et al. Determination of a broad structural ensemble representing the denatured state of the bovine acyl-coenzyme A binding protein. J. Am. Chem. Soc. 126, 3291–3299 (2004)
Korzhnev, D. M. et al. Low-populated folding intermediates of Fyn SH3 characterized by relaxation dispersion NMR. Nature 430, 586–590 (2004)
Vendruscolo, M., Paci, E., Dobson, C. M. & Karplus, M. Three key residues form a critical contact network in a protein folding transition state. Nature 409, 641–645 (2001)
Lindorff-Larsen, K., Paci, E., Vendruscolo, M. & Dobson, C. M. Transition states for protein folding have native topologies despite high structural variability. Nature Struct. Mol. Biol. 11, 443–449 (2004)
Schwieters, C. D. & Clore, G. M. Reweighted atomic densities to represent ensembles of NMR structures. J. Biomol. NMR 23, 221–225 (2002)
Jorgensen, W. J., Chandrasekhar, J., Madura, J. D., Impey, R. W. & Klein, M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983)
Brooks, B. R. et al. CHARMM: A program for macromolecular energy, minimization and dynamics calculations. J. Comput. Chem. 4, 187–217 (1983)
Paci, E. & Karplus, M. Forced unfolding of fibronectin type 3 modules: An analysis by biased molecular dynamics simulations. J. Mol. Biol. 288, 441–459 (1999)
Fox, T. & Kollman, P. A. The application of different solvation and electrostatic models in molecular dynamics simulations of ubiquitin: How well is the X-ray structure ‘maintained’? Proteins 25, 315–334 (1995)
Acknowledgements
We are very grateful to P. I. de Bakker and T. L. Blundell for assistance in determining the X-ray rapper ensemble of ubiquitin. We thank S. E. Jackson for sharing the experimental data on ubiquitin stability changes before publication. K.L.L. is supported by the Danish Research Agency. M.A.D. was funded by the Marshall Aid Commemoration Commission, US National Science Foundation, and Cambridge Overseas Trust. M.V. is a Royal Society University Research Fellow. The research of M.V. and C.M.D. is supported in part by Programme Grants from the Wellcome and Leverhulme Trusts.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Supplementary information
Supplementary Methods
Details on all methods for structure determination and analysis. (PDF 62 kb)
Rights and permissions
About this article
Cite this article
Lindorff-Larsen, K., Best, R., DePristo, M. et al. Simultaneous determination of protein structure and dynamics. Nature 433, 128–132 (2005). https://doi.org/10.1038/nature03199
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature03199