Abstract
There are at least three RNA silencing pathways for silencing specific genes in plants. In these pathways, silencing signals can be amplified and transmitted between cells, and may even be self-regulated by feedback mechanisms. Diverse biological roles of these pathways have been established, including defence against viruses, regulation of gene expression and the condensation of chromatin into heterochromatin. We are now in a good position to investigate the full extent of this functional diversity in genetic and epigenetic mechanisms of genome control.
Similar content being viewed by others
Main
Although RNA silencing has only emerged as a topic of general interest in the past six years, the first RNA silencing paper may have been published as long ago as 1928. In that paper Wingard described tobacco plants in which only the initially infected leaves were necrotic and diseased owing to tobacco ringspot virus1 (Fig. 1). The upper leaves had somehow become immune to the virus and consequently were asymptomatic and resistant to secondary infection. At the time this ‘recovery’ was a mystery: there was no obvious way to explain the specificity of the resistance to secondary infection.
The original legend1 to the figure reads ‘Turkish tobacco plant 23 days after inoculation with ringspot. Note the gradual decline in the development of ringspot symptoms on the upper leaves until finally the top leaves appear perfectly normal’. We now know that the virus causing the initial symptoms had activated viral RNA silencing that inhibited spread of the infection into the upper leaves, and caused them to be specifically immune to tobacco ringspot virus secondary infection.
The details of the tobacco ringspot virus example remain to be worked out but we now know that recovery from virus disease involves RNA silencing that is targeted specifically at the viral RNA2,3. There was no information about mechanisms in 1928 — it was not even known that the viral genome is RNA. But Wingard's paper is an appropriate starting point for the current interest in RNA silencing because it illustrates a viral defence role for RNA silencing which may have been one of its original functions in primitive eukaryotes. In modern plants this process has diversified into mechanisms that, in addition to defending the plant against viruses, protect the genome from transposons and regulate gene expression.
Here, I describe three natural pathways of RNA silencing in plants that have been revealed by genetic and molecular analysis. These pathways all involve the cleavage of a double-stranded RNA (dsRNA) into short 21–26-nucleotide RNAs by an enzyme Dicer that has RNase III domains. These RNAs are known as short interfering RNAs (siRNAs) and microRNAs (miRNAs). I discuss the possibility that there may be more than these three pathways, or that several variant mechanisms of RNA silencing exist. I also discuss features that distinguish plant miRNAs from animal miRNAs, and the amplification and mobile signal mechanisms in the siRNA pathways. Finally, I speculate about the role of RNA silencing in the integration of genome regulation.
Diverse RNA silencing pathways
The first pathway of the three is cytoplasmic siRNA silencing4. This pathway may be important in virus-infected plant cells where the dsRNA could be a replication intermediate or a secondary-structure feature of single-stranded viral RNA. In the case of plant DNA viruses, the dsRNA may be formed by annealing of overlapping complementary transcripts. The originally described recovery from tobacco ringspot virus1 and many examples of transgene silencing in plants and animals are probably manifestations of cytoplasmic RNA silencing.
The second pathway is the silencing of endogenous messenger RNAs by miRNAs. These miRNAs negatively regulate gene expression by base pairing to specific mRNAs, resulting in either RNA cleavage or arrest of protein translation. Like siRNAs, the miRNAs are short 21–24-nucleotide RNAs derived by Dicer cleavage of a precursor. The prototype miRNAs in plants were identified as a subset of the short RNA population, with the molecular characteristics of the heterochronic RNAs let-7 and lin-4 in Caenorhabditis elegans5. miRNAs are derived from an inverted repeat precursor RNA with partially double-stranded regions, and they target a complementary single-stranded mRNA. However, there are differences between the miRNAs of plants and animals, as discussed in section ‘miRNA in plants’ below (see review in this issue by Ambros, page 350, for a more complete discussion of miRNAs in animals).
The third pathway of RNA silencing in plants is associated with DNA methylation and suppression of transcription. The first evidence for this type of silencing was the discovery in plants that transgene and viral RNAs guide DNA methylation6,7,8 to specific nucleotide sequences. More recently, these findings have been extended by the observations that siRNA-directed DNA methylation in plants is linked to histone modification9, and that, in fission yeast, hetero-chromatin formation at centromere boundaries is associated with siRNAs10. An important role of RNA silencing at the chromatin level is probably protecting the genome against damage caused by transposons (see review in this issue by Lippman and Martienssen, page 364).
An ancient origin of these three pathways of RNA silencing is likely because there are examples of each type in animals, fungi and plants. Green plants are unusual in that they have retained the capacity for all three types of silencing, whereas other organisms may have lost one or more of these pathways. Budding yeast, for example, has apparently lost all RNA silencing, and in mammals, all the examples of natural silencing found so far involve miRNAs5. Although exogenous RNAs in mammalian cells initiate ‘classical’ RNA interference (RNAi; a type of RNA silencing) involving siRNAs, it is not clear whether a specific siRNA pathway is involved. It could be that the exogenous RNAs are recruited into the miRNA pathway.
Argonaute and Dicer gene families
The Argonaute (Ago) proteins in plants, animals and fungi have been implicated in all three pathways of RNA silencing. In Arabidopsis thaliana, for example, AGO1 mutants are defective for cytoplasmic RNA and miRNA silencing pathways, and AGO4 mutants are impaired in chromatin silencing9. A central role of these AGO proteins seems likely because they are components of the silencing effector complexes that bind to siRNAs and miRNAs. Thus, the Drosophila melanogaster AGO2 protein binds siRNA by means of the PAZ (for piwi–argonaute–zwille) domain11, and is in the ribonuclease complex RISC (RNA-induced silencing complex)12 that cleaves the target mRNA. Its role in RISC, on the basis of evidence with mouse AGO2 protein, is probably the ‘slicer’ ribonuclease in RISC13. AGO1 is probably a RISC component in A. thaliana because hypomorphic mutants retain the ability to accumulate miRNA, but the corresponding target mRNAs are not cleaved14. In fission yeast, an Ago protein is found in the RNA-induced transcriptional silencing complex (RITS) that targets heterochromatinization15. Finally, in Tetrahymena, an Ago homologue and siRNAs are found in a complex implicated in a silencing-related mechanism that leads to genome rearrangement16,17.
This role in silencing effector complexes indicates that all silencing mechanisms will involve an Ago protein. Conversely it is likely that many if not all Ago proteins will be silencing-related. If this is the case, at least some of the ten Ago homologues in the A. thaliana genome may be associated with effector complexes of RNA silencing. Perhaps they associate with RISC or RITS that are adapted to silence genes in specialized cells, or at particular developmental stages. The mutant phenotype for AGO7 in A. thaliana, for example, is altered timing of the phase change between juvenile and adult leaves: so AGO7 might be part of an RNA silencing effector complex which is specific to a particular stage of development18.
The diverse RNA silencing pathways may not be completely separate. Some proteins, for example AGO1 and zwille (AGO10), have partially overlapping functions. Conversely, as illustrated by the miRNA- and cytoplasmic-siRNA-defective phenotype of ago1 mutants in A. thaliana14,19, a single Ago protein may participate in multiple silencing pathways. Mutations at another silencing-related gene, HEN1, also indicate overlap in different silencing pathways: HEN1 mutants are defective in both miRNA and cytoplasmic siRNA silencing20.
The Dicer gene family in A. thaliana has only four members21: presumably, if there are more than four silencing pathways involving Ago proteins, some Dicers will be active in more than one of them. Two Dicer proteins in A. thaliana have well defined functions: Dicer-like (DCL)1 is required for miRNA biogenesis22,23,24; DCL3 produces retroelement and transposon siRNAs and is required for chromatin silencing24. The DCL3 products correspond to a class of siRNA that, from earlier analyses, had been associated with transposons25, and that are longer (24 nucleotides rather than 21 nucleotides)25,26 than the typical DCL1 products. However the role of the other two Dicers, DCL2 and DCL4, has been more difficult to define. DCL2 has been implicated in viral siRNA production but the loss-of-function phenotype is only a transient reduction in the level of siRNA in one of several viruses tested24. It is likely, therefore, that there is functional redundancy and that the other Dicers of A. thaliana are also involved in viral siRNA production. The function of DCL4 is not known.
miRNAs in plants
Plant and metazoan miRNA pathways are fundamentally the same: both involve 20–22-nucleotide single-stranded miRNAs that are generated by a Dicer and both depend on an Ago protein. However, the metazoan miRNAs are processed by Drosha and Dicer RNase III in two steps that take place in the nucleus and cytoplasm5, whereas miRNAs in plants are processed by a Dicer, and this is most likely to occur in the nucleus22. Associated with this processing difference, a dsRNA binding protein, HYL1, is specific to the plant miRNA pathway 27,28. A further important difference is that the plant miRNAs are more perfectly paired to their target RNA and use RNA cleavage rather than translation suppression as the primary silencing mechanism29,30,31. The animal miRNAs are normally targeted to the 3′ untranslated region (UTR) of a mRNA, whereas the plant miRNAs have targets in the coding sequence or even in the 5′ UTR32.
There are now extensive lists of plant miRNAs (http://cgrb.orst.edu/smallRNA/ and http://www.sanger.ac.uk/Software/Rfam/mirna/) and, in several cases, the target mRNA has been validated experimentally by expression of an miRNA-resistant target gene with silent mutations in the putative miRNA complementary region (Fig. 2). The mutations interfere with miRNA targeting and result in overexpression of a bona fide miRNA target. This gold standard of miRNA target identification has been applied to target mRNAs encoding: (1) a TCP transcription factor required for leaf morphogenesis33; (2) a MYB33 transcription factor involved in a plant hormone response33; (3) an HD-ZIP transcription factor that influences abaxial and adaxial polarity in leaves and stems34; (4) the AP2 transcription factor that regulates floral development35; (5) a NAC transcription factor that plays a role at many stages of development36; and (6) the AGO1 cofactor of silencing14. Other examples that have been validated by the detection of target mRNA cleavage products corresponding to an miRNA-complementary site include a scarecrow transcription factor of unknown function37 and DCL1 (ref. 38).
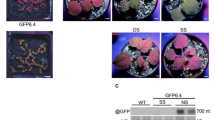
In a wild-type plant (a) an miRNA associated with RISC will base pair to its cognate target and promote either sequence-specific RNA degradation or a translational block. However, if a transgene is introduced in which the miRNA target sequence has silent mutations (b), the miRNA cannot bind to the target sequence and the protein encoded by the mRNA is overexpressed. c, Cross-sections of stems in wild-type and rev-10d transgenic A. thaliana, illustrating a phenotype from a miRNA-resistant mRNA (from ref. 34). The rev10d transgene encodes the revoluta transcription factor and its RNA is resistant to targeting by miR165 and miR166. In the wild-type plants the xylem (xy) is positioned centrally and inside the peripheral phloem (ph) tissue. In the rev-10d stems the vascular bundles are radialized with xylem tissue (arrowheads) surrounding phloem tissue (ph). This effect on the distribution of xylem tissue implies that interpretation of positional information requires correct targeting by miR165 and miR166. 7mG, 7-methylguanine.
These, and many other of the miRNAs in A. thaliana for which the targets have been identified computationally, correspond to mRNAs for transcription factors and other proteins involved in developmental regulation30,31,39. The mRNAs for proteins associated with ubiquitin-mediated protein degradation are also potential miRNA targets30,32. The target sequences in most of these examples are conserved in rice and A. thaliana and, in one instance, the conservation extends even between mosses and flowering plants40. However, the range of targets is not restricted to ‘developmental’ genes because there are also miRNAs that increase or decrease in abundance following cold or drought stress or sulphur starvation. The predicted targets of these miRNAs encode a more diverse set of proteins including laccases, cytochrome c oxidases, spliceosomal proteins and ATP sulphurylases30,32.
It has been estimated that the A. thaliana genome has about 100 miRNA loci30. This estimate, however, is based on a computational genome survey which assumes that miRNA targets are conserved in A. thaliana and rice. Putative miRNA targets that are conserved between A. thaliana and Lotus, Medicago or Populus and not rice32, or that are not conserved in distantly related species, would not have been identified in this survey and the number of miRNA loci could be considerably higher.
Initiation and amplification of silencing
RNA-dependent RNA polymerases (RDRs; also known as RdRPs) are required for the cytoplasmic and chromatin RNA silencing pathways in C. elegans41,42, fungi10,43 and plants24,44,45 but not, apparently, for the same pathways in insects or mammals. The RDRs share a common sequence motif that is distantly related to the catalytic domain of DNA-dependent RNA polymerases46, and it is therefore likely that they are an ancient group of proteins. A. thaliana, C. elegans and Neurospora crassa have small RDR gene families that, as with Ago proteins, indicate functional diversification of silencing pathways. In A. thaliana the RDR1 and RDR6 (also known as SDE1/SGS2) orthologues are required in the cytoplasmic RNA silencing pathway that silences transgenes and viruses. However, it seems that these proteins have specificity for different viral RNAs: RDR6 mutants in A. thaliana are hypersusceptible to cucumber mosaic virus45 but not to tobacco rattle and tobacco mosaic virus47, whereas tobacco plants with reduced levels of RDR1 show enhanced susceptibility to tobacco mosaic virus48. RDR2 mutants are defective for production of endogenous siRNAs, including those corresponding to retroelements. So the RDR2 protein might be part of the chromatin silencing pathway24.
In principle, the RDR proteins could mediate primer-dependent and primer-independent mechanisms of RNA silencing (Fig. 3). The primer-independent process may be important for the production of dsRNA from a single-stranded template, so that silencing can be initiated in virus-infected plants or with transgene RNAs (Fig. 3a). Consistent with this primer-independent mechanism, in vitro assays with N. crassa49 and tomato enzymes50 demonstrate that RDR catalyses primer-independent synthesis of dsRNA on a single-stranded RNA (ssRNA) template. Similarly, in wheat germ extracts, ssRNA can be copied into complementary RNA by an unidentified enzyme that, presumably, is an RDR26. However, it is not yet clear how RDR could differentiate the viral and transgene RNAs targeted for silencing from the non-silenced endogenous RNAs. Perhaps the RNA that becomes silenced contains ‘aberrant’ features that are absent from ‘normal’ non-silenced RNA. Alternatively, the aberrant RNA might lack features that are present in normal RNA. The absence of a 5′ cap (R. Sablowski, personal communication) renders an RNA susceptible to RDR-dependent RNA silencing in A. thaliana, but other possibilities have not been ruled out.
a, RNAs are normally not silenced because the RDR proteins do not have access to the template RNA sequence. Cap-binding protein (CBP) and poly-adenosine-binding protein (PABP) may be involved in this restriction of RDR access. However in b, the RDR protein is allowed access because the RNA lacks a 5′ cap or 3′ poly-adenosine tail, and dsRNA is produced which enters the siRNA pathway. b, The amplification process would result from the ability of a single aberrant RNA to generate many molecules of siRNA. c shows the outcome if a small quantity of primary siRNA is present from either a virus, a transposon or from a cellular RNA through the process shown in b. The antisense strand of this siRNA may anneal by base pairing to a target RNA and serve as a primer for the RDR. The resulting dsRNA would then be cleaved by Dicer and, as in b, there would be amplification because many secondary siRNAs would be produced from each molecule of primary siRNA.
The second RDR mechanism (Fig. 3b) requires that primary siRNAs from a virus, transposon or transgene are primers in RDR-directed synthesis of dsRNA. The QDE1 RDR protein from N. crassa incorporates a labelled 20-nucleotide antisense RNA into the complementary strand of a ssRNA in vitro49, in a manner that is consistent with this mechanism. This primer-dependent process is also supported by indirect genetic evidence from C. elegans and plant systems in which the initiator of silencing comes from part of a target gene. In these systems, the secondary siRNAs that accumulate in the silenced tissue are dependent on RDR proteins41,51, and are derived not only from the initiator region but also from adjacent regions in the target sequence. Moreover, the secondary siRNAs in C. elegans correspond only to the 5′ side of the ssRNA, as would be expected if an antisense primary siRNA had been extended by the RDR at its 3′ end. However, in A. thaliana and Nicotiana benthamiana the secondary siRNAs are from both the 5′ and the 3′ side of the initiator51,52 on the ssRNA, and so cannot be produced from a simple priming mechanism on a single RNA species. The most likely explanation here is that the silencing target, like many parts of the A. thaliana genome, is transcribed from both strands53. The 3′ secondary siRNAs would then result from extension of an siRNA primer on an antisense RNA template.
As a result of the RDR-mediated mechanisms, a single aberrant RNA species or primary siRNA molecule could generate many dsRNAs which would then silence even more target molecules. This amplification process is likely to be essential in virus defence because it would ensure that silencing of viral RNAs keeps pace with the replication and accumulation of viral RNA. Similarly, in genome defence, the amplification steps would ensure that a few molecules of transposon RNA could activate the chromatin-silencing pathway sufficiently to suppress all copies of a transposable element. In addition, the RDR proteins would help target the RNA silencing mechanism to transposons because transcripts with direct repeats are readily amplified54 (see review in this issue by Lippman and Martienssen, page 364).
Mobile silencing signals
Together with RDR amplification, mobility of a silencing signal is probably a crucial characteristic of an antiviral defence system. A mobile silencing signal could move either with or ahead of the virus to silence the viral RNA before, or at the same time, as the virus moves into a cell55. Indeed, in plants and C. elegans, the effects of silencing extend beyond those cells in which the silencing is initiated and can spread systemically56,57,58 through the organism. This systemic effect has nucleotide-sequence specificity corresponding to the initiator dsRNA, indicating that the signal either is an RNA or that it has an RNA component. Consistent with this idea, the SID1 protein, which is required for systemic RNAi in C. elegans, is a transporter of dsRNA across membranes59,60.
In plants the systemic silencing mechanism is unlikely to be the same as that in C. elegans. The signal does not have to cross any membranes because most of the cells in a plant, including the phloem cells of the vascular system, are connected by plasmodesmatal channels that are a continuation of the endoplasmic reticulum61. So far, none of the host proteins involved in movement of this silencing signal has been identified. However, an analysis of systemic signalling from a green fluorescent protein (GFP) transgene coupled to a phloem-specific promoter indicated that the signalling mechanism in plants can be resolved into short (up to 15 cells) and longer range62 phases extending up to several centimetres.
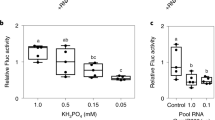
Short-range signalling is unlike the longer range movement because it is unaffected by RDR6 loss-of-function mutants (Fig. 4), and it is likely that a 21-nucleotide siRNA is the mobile signal62. Consistent with a short RNA being the mobile signal for short-range signalling, the siRNA in a virus-infected cell is present either as free RNA or in low molecular weight complexes that could be well below the normal size exclusion limit of plasmodesmata63.
The two panels show A. thaliana plants carrying two transgenes. a, In the wild-type plant, a GFP transgene is constitutively transcribed, but the GFP fluorescence is suppressed because a second transgene for GFP dsRNA is expressed in the phloem cells. A silencing signal has moved out of the phloem and has silenced the GFP transgene throughout the leaf. The plant appears red under ultraviolet light owing to chlorophyll fluorescence. b, This plant has a mutation in RDR6 and the silencer signal is able to act only in the cells that are close to the phloem. The GFP transgene is not silenced in cells that are further than about 20 cells from the phloem and consequently they appear green under ultraviolet light (reproduced with permission from ref. 62.).
A longer 24-nucleotide class of siRNAs, possibly generated by DCL3 (see section ‘Argonaute and Dicer gene families’ above), has been proposed as a candidate for the long-range phloem entry signal because viral proteins that block systemic silencing also prevent accumulation of the 24-nucleotide siRNA25. However, systemic silencing is transmitted from grafted plants in which both the 21- and 24-nucleotide siRNAs are suppressed by the viral HCPro suppressor of silencing64. It is therefore possible that other silencing RNAs including long ssRNA, dsRNA or siRNAs, could be signal molecules because any of them can initiate silencing if they are introduced into a cell with a suitable target. The plasmodesmatal size exclusion limit61 might be a barrier to intercellular movement of high molecular weight complexes containing these RNAs, but it is possible that free RNAs or RNAs in low molecular weight complexes are mobile. Precedents for phloem-mobile RNAs in plants include viroids65 — small non-protein coding RNA plant pathogens — and the transcript corresponding to a homeobox protein mRNA66.
An intriguing possibility is that the movement of miRNAs and endogenous siRNAs might play a role in regulation of endogenous genes. For example, during leaf development miR165 and miR166 are negative regulators of genes affecting leaf polarity, and their distribution and possible gradient of expression is consistent with that of a mobile signal34,67,68. Consistent with the idea that endogenous silencing RNAs are mobile signals, there are many miRNAs and siRNAs in the phloem sap of pumpkin69, and a phloem protein has been detected that binds specifically to the single-stranded form of these RNAs.
Viral suppressor proteins
RNA silencing in plants prevents virus accumulation and, accordingly, viruses have evolved various strategies to counteract this defence mechanism. The primary counter defence measure involves suppressor proteins of silencing which are encoded in the genomes of both RNA and DNA viruses70. These proteins probably evolved independently in different virus groups because they are structurally diverse, and there are no common sequence motifs. A secondary mechanism to counteract silencing is illustrated by the apparent resistance of satellite and defective interfering RNAs to degradation by siRNAs71,72. It seems that these RNAs have protective secondary structures, or are compartmentalized so that they are hidden from the RNA silencing mechanism.
In principle, the plant viral suppressor proteins could be used as a tool to investigate the mechanism of RNA silencing. However, in most instances, including the prototype HCPro suppressor from potyviruses, there are conflicting data about their mechanism of action73,74,75. Only for two suppressors — p21 encoded by beet western yellow virus73 and p19 encoded by the tomato bushy stunt virus (TBSV) group63,74 — is there a clear indication of how they act. In both cases, the virus suppressor protein binds to and, presumably, inactivates siRNAs so that they do not target the corresponding viral RNAs. For p19, the high resolution crystal structures for two different TBSV-group proteins, combined with molecular and biochemical data, indicate precisely how silencing is blocked. A tail-to-tail p19 homodimer forms α-helix brackets around the ends of the siRNA base-paired region76,77 and, consequently, an siRNA or miRNA is prevented from being incorporated into an active RISC63,74,78. In transgenic Arabidopsis expressing p19 (ref. 73), both miRNA and its complement (miRNA*) accumulate, whereas in the control plants without p19 the miRNA* is undetectable74. Presumably the miRNA–miRNA* duplex is normally a short-lived precursor of miRNA–RISC79, but is stabilized in the presence of p19.
Given the likelihood that virus defence was an ancient role of RNA silencing, it would not be surprising if RNA silencing also influences animal virus infections. Consistent with this idea, the NS1 and E3L proteins of influenza and vaccinia viruses78, and the B2 protein of flock house virus80, have silencing suppressor activity. In addition, there are five different miRNAs in mammalian cells infected with Epstein–Barr virus that correspond to inverted repeat regions in the viral genome81. One of these viral miRNAs targets the viral DNA polymerase gene BALF5, and directs processing of a BALF5 trancript which is likely to affect virus accumulation. However, this is the only report of siRNAs or miRNAs corresponding to mammalian viruses, and it remains to be confirmed whether the silencing suppressor activity of the influenza and vaccinia virus proteins is a side effect of their dsRNA binding activity82. So, on the basis of current evidence, it seems unlikely that RNA silencing in mammals is a general defence mechanism against viruses as it is in plants. Perhaps it is effective against a subset of mammalian viruses or is an antiviral defence in embryonic or other cells in which the systems of innate and humoral immunity are ineffective.
Viral symptoms and silencing
As viruses are inducers, suppressors and targets of the RNA silencing mechanism, there are many ways in which the symptoms in infected plants can be influenced by viral intervention in the miRNA and siRNA pathways. For example, in A. thaliana plants infected with turnip mosaic virus, the symptoms include developmental defects that are mimicked by transgenic expression of the HCPro suppressor of silencing, or by mutation affecting the DCL1 protein of the miRNA pathway83. It is likely, therefore, that these symptoms are caused by suppression of the host's miRNA pathway by the viral HCPro. By extrapolation, other viral symptoms involving developmental defects are probably due to silencing suppressors33,34,68.
A variation on this effect of viral suppressors is suggested by the finding that transgenic tobacco plants expressing HCPro show enhanced resistance to diverse pathogens including tobacco mosaic virus and the oomycete (water mould) Perenospora tabacina. One plausible explanation for this resistance is that HCPro suppresses the action of endogenous miRNAs or siRNAs that usually target negative regulators of the host's innate immune system84.
A second more direct role of RNA silencing in symptom formation is illustrated by cucumber mosaic virus strains with small noncoding satellite RNAs. The Y strain of satellite RNA causes a bright yellow chlorosis in infected plants72 that is suppressed in transgenic plants expressing the HCPro silencing suppressor. Presumably, the symptoms develop because there are Y-satellite-RNA-derived siRNAs that target an endogenous gene normally required for chlorophyll accumulation. Similarly in viroid infections, pathogen-derived siRNAs may target endogenous genes72. Viroid RNAs would be an excellent substrate for Dicer because they are essentially circular RNAs that fold to form an extensively double-stranded rod-like structure. Consistent with the involvement of siRNAs, viroid symptoms are blocked if the infected plant expresses HCPro, and are mimicked in transgenic plants expressing an inverted repeat transgene derived from the symptom-inducing region of the viroid genome72.
RNA silencing in genetic regulation
One of the challenges for research in genetic regulation is to understand how expression of whole sets of genes can be coordinated. RNA silencing may contribute to this integration of genetic control because it is subject to several levels of feedback regulation, and because any one siRNA or miRNA could target the silencing mechanism to multiple RNAs or to DNA loci. The combined effect of these features could be that a single RNA species mediates RNA silencing-based effects on many other genes and RNAs. One level of feedback control is illustrated by cytoplasmic RNA silencing in which dsRNA is generated from a ssRNA by an RDR (Fig. 3a). In this scenario the original ssRNA template is both a target and a precursor of the siRNA. High levels of the ssRNA would therefore lead to abundant siRNA and, consequently, the ssRNA levels would decline. Conversely, reduced levels of the ssRNA would lead to decreased amounts of siRNA and ultimately an increase in the amount of ssRNA (Fig. 5a).
a, The sequence of events when siRNA production involves an RDR using a ssRNA template. The siRNA is incorporated into RISC and negatively regulates its own production by targeting RISC at the ssRNA. b, The feedback inhibition of miR162 on its target mRNA encoding DCL1 Dicer (ref. 38). DCL1 mediates the production of miR162 from the pre-miR162 precursor RNA. The miR162 then targets the DCL1 mRNA, and negatively regulates DCL1 synthesis. So a high level of miR162 leads to a decrease in the rate of DCL1 production, whereas a low level of miR162 has the opposite effect. c, A negative-feedback regulatory mechanism proposed for miR159. The target RNA of miR159 encodes a MYB33 transcription factor. Both MYB33 mRNA and miR159 are positively regulated by the plant hormone GA85: in a GA-responding A. thaliana there is an increase in the level of the MYB33 mRNA which is associated with an increase in the level of miR159. The high levels of miR159 then suppress the GA-stimulated increase in MYB33. Several rounds of the priming process would amplify the silencing effect of the siRNA.
Feedback mechanisms are also apparent in the miRNA pathways because the mRNA transcripts encoding DCL1 (ref. 38) and the AGO1 component of RISC14 are themselves targets of miRNAs (miR162 and miR168, respectively). Abundant AGO1 or DCL1 proteins would lead to a silencing-mediated decrease in the amount of corresponding mRNAs. Conversely, reduced amounts of these proteins would ease the level of silencing and the concentration of mRNAs would increase. This feedback mechanism could explain the otherwise paradoxical increase in the amount of miRNA in the presence of viral suppressors of silencing73,74,75 (Fig. 5b). In this case, the suppression of silencing would uncouple the feedback loop so that the abundance of AGO1, DCL1 and the associated miRNAs would be unchecked by the normal mechanisms.
A second type of feedback control is implied by the finding that miR159 and its putative target (transcription factor MYB33 mRNA) are both positively regulated by the plant hormone gibberellic acid (GA)85. A GA stimulus could lead to an increase in MYB33 that would initiate flowering and, directly or indirectly, to an increase in miR159. The higher concentration of miR159 would then counteract the increase in MYB33 and dampen the GA response (Fig. 5c). A similar mechanism may apply to miR171 that targets the transcription factor GRAS mRNA31 . If miR171 were a simple negative regulator of the transcription factor mRNA then the miRNA would be abundant when the target is rare and vice versa. In fact both are upregulated in inflorescences31 in a pattern that could be explained if the GRAS transcription factor promoted expression of the miR171.
The ability of siRNAs and miRNAs to target multiple RNAs and DNAs may also contribute to their role in integration of genetic regulation. Targeting by these short RNAs is tolerant of limited sequence mismatches5, and each siRNA or miRNA probably has both primary and secondary targets. The primary targets would be more completely matched to the siRNA or miRNA and, if the target is an RNA, it would be cleaved by RISC. Any secondary targets would have more mismatches and would be cleaved more slowly than the primary targets. Alternatively, as occurs for APETALA2 (AP2) mRNA35,86, the secondary RNA targets might be translationally repressed rather than degraded. Currently there is no information about secondary targets of siRNAs and miRNAs in plants. Investigation of miR159 and miR319 may tell us about the potential importance of secondary targets because the two miRNAs differ at only three nucleotide positions and have MYB or TCP mRNAs, respectively, as distinct primary targets33. The extent to which these miRNAs cross target at either the RNA cleavage or translational repression level will indicate the extent to which miRNAs in plants might have multiple targets.
Future prospects
Over the past few years, we have come to appreciate that there are diverse natural roles of RNA silencing, ranging from defence against viruses to the regulation of gene expression and chromosome structure. But a remaining challenge is to find out the full extent of this functional diversity. One approach will be to analyse Ago and other gene families associated with the silencing mechanism. The characterization of silencing-related RNAs using computational approaches and direct sequence analysis may also be informative, as is discussed above in the context of miRNAs. In addition to miRNAs, plants including A. thaliana have vast numbers of other short RNA species, many of which are likely to be siRNAs23,28. A small RNA database (http://cgrb.orst.edu/smallRNA/) lists more than 2,000 of them but there are likely to be many more, and their extended analysis will probably be a rich source of information about genetic elements that are targeted by RNA silencing.
The RNA targeting mechanisms of RNA silencing have not been extensively investigated in plants: one particularly neglected area is the possibility of translational suppression, as is mediated by animal miRNAs5. One plant miRNA (miR172) is known to suppress translation of the floral regulator AP2 mRNA35,86, but for most other miRNAs and siRNAs the possibility of translational effects has not been investigated. Transgene silencing87 suggests that translational effects may be more general, and perhaps protein rather than RNA profiling should be used to identify the targets of RNA silencing mechanisms.
Another outstanding question concerns silencing RNAs that move between cells. Current indications are that this signalling process affects virus movement55 (E. Bayne, F. Schwach and D.B., unpublished work), but possible roles in plant growth and development have yet to be explored. miR165 and miR166 regulate spatial information in plant development34,67 and conceivably the movement of silencing-related RNAs is involved (Fig. 2).
Although RNA silencing is a relatively recent topic of research, the mechanisms are now becoming clear and there is some indication of its biological roles. The discovery of RNA silencing has completely changed our view of RNA as a regulatory molecule in eukaryotic cells and it is likely that this view will continue to evolve as further discoveries emerge about the diversity of silencing mechanisms.
References
Wingard, S. A. Hosts and symptoms of ring spot, a virus disease of plants. J. Agric. Res. 37, 127–153 (1928).
Ratcliff, F., Harrison, B. D. & Baulcombe, D. C. A similarity between viral defense and gene silencing in plants. Science 276, 1558–1560 (1997).
Covey, S. N., Al-Kaff, N. S., Langara, A. & Turner, D. S. Plants combat infection by gene silencing. Nature 385, 781–782 (1997).
Hamilton, A. J. & Baulcombe, D. C. A species of small antisense RNA in post-transcriptional gene silencing in plants. Science 286, 950–952 (1999).
Bartel, D. P., MicroRNAs: genomics biogenesis, mechanism, and function. Cell 116, 281–297 (2004).
Wassenegger, M., Heimes, S., Riedel, L. & Sanger, H. L. RNA-directed de novo methylation of genomic sequences in plants. Cell 76, 567–576 (1994).
Mette, M. F., Aufsatz, W., van der Winden, J., Matzke, M. A. & Matzke, A. J. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 19, 5194–5201 (2000).
Jones, L., Ratcliff, F. & Baulcombe, D. C. RNA-directed transcriptional gene silencing in plants can be inherited independently of the RNA trigger and requires Met1 for maintenance. Curr. Biol. 11, 747–757 (2001).
Zilberman, D., Cao, X. & Jacobsen, S. E. ARGONAUTE4 control of locus specific siRNA accumulation and DNA and histone methylation. Science 299, 716–719 (2003).
Volpe, T. et al. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297, 1833–1837 (2002).
Lingel, A., Simon, B., Izaurralde, E. & Sattler, M. Structure and nucleic-acid binding of the Drososphila Argonaute2 PAZ domain. Nature 426, 465–469 (2003).
Hammond, S. M., Boettcher, S., Caudy, A. A., Kobayashi, R. & Hannon, G. J. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293, 1146–1150 (2001).
Liu, J. et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science published online 29 July 2004 (doi:10.11261/science.1102513).
Vaucheret, H., Vazquez, F., Crete, P. & Bartel, D. P. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 18, 1187–1197 (2004).
Verdel, A. et al. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303, 672–676 (2004).
Taverna, S. D., Coyne, R. S. & Allis, D. C. Methylation of histone H3 at lysine 9 targets programmed DNA elimination in Tetrahymena. Cell 110, 701–711 (2002).
Mochizuki, K., Fine, N. A., Fujisawa, T. & Gorovsky, M. A. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in Tetrahymena. Cell 110, 689–699 (2002).
Hunter, C., Sun, H. & Poethig, R. S. The Arabidopsis heterochronic gene ZIPPY is an ARGONAUTE family member. Curr. Biol. 13, 1734–1739 (2003).
Fagard, M., Boutet, S., Morel, J.-B., Bellini, C. & Vaucheret, H. AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc. Natl Acad. Sci. USA 97, 11650–11654 (2000).
Boutet, S. et al. Arabidopsis HEN1: A genetic link between endogenous miRNA controlling development and siRNA controlling transgene silencing and virus resistance. Curr. Biol. 13, 843–848 (2003).
Schauer, S. E., Jacobsen, S. E., Meinke, D. W. & Ray, A. DICER-LIKE1: blind men and elephants in Arabidopsis development. Trends Plant Sci. 7, 487–491 (2002).
Papp, I. et al. Evidence for nuclear processing of plant microRNA and short interfering RNA precursors. Plant Physiol. 132, 1382–1390 (2003).
Finnegan, E. J., Margis, R. & Waterhouse, P. M. Posttranscriptional gene silencing is not compromised in the Arabidopsis CARPEL FACTORY (DICER-LIKE1) mutant, a homolog of Dicer-1 from Drosophila. Curr. Biol. 13, 236–240 (2003).
Xie, Z. et al. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2, E104 (2004).
Hamilton, A. J., Voinnet, O., Chappell, L. & Baulcombe, D. C. Two classes of short interfering RNA in RNA silencing. EMBO J. 21, 4671–4679 (2002).
Tang, G., Reinhart, B. J., Bartel, D. P. & Zamore, P. D. A biochemical framework for RNA silencing in plants. Genes Dev. 17, 49–63 (2002).
Han, M.-H., Goud, S., Song, L. & Fedoroff, N. The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc. Natl Acad. Sci. USA 101, 1093–1098 (2004).
Vazquez, F., Gasciolli, V., Crete, P. & Vaucheret, H. The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr. Biol. 14, 346–351 (2004).
Rhoades, M. W. et al. Prediction of plant microRNA targets. Cell 110, 513–520 (2002).
Jones-Rhoades, M. W. & Bartel, D. P. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell 14, 787–799 (2004).
Llave, C., Kasschau, K. D., Rector, M. A. & Carrington, J. C. Endogenous and silencing-associated small RNAs in plants. Plant Cell 14, 1605–1619 (2002).
Sunkar, R. & Zhu, J.-K. Novel and stress-regulated miRNAs and other small RNAs from Arabidopsis. Plant Cell (in the press).
Palatanik, J. F. et al. Control of leaf morphogenesis by microRNAs. Nature 425, 257–263 (2003).
Emery, J., Floyd, S. K., Alvarez, J., Baum, S. F. & Bowman, J. L. Radial patterning of Arabidopsis shoots by Class III HD-ZIP and KANADI genes. Curr. Biol. 13, 1768–1774 (2003).
Chen, X. M. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303, 2022–2025 (2004).
Mallory, A. C., Dugas, D. V., Bartel, D. P. & Bartel, B. MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr. Biol. 14, 1035–1046 (2004).
Llave, C., Xie, Z., Kasschau, K. D. & Carrington, J. C. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297, 2053–2056 (2002).
Xie, Z., Kasschau, K. D. & Carrington, J. C. Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr. Biol. 13, 784–789 (2003).
Park, W., Li, J., Song, R., Messing, J. & Chen, X. CARPEL FACTORY: a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12, 1484–1495 (2002).
Floyd, S. K. & Bowman, J. L. Gene regulation: ancient microRNA target sequences in plants. Nature 428, 485–486 (2004).
Sijen, T. et al. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107, 465–476 (2001).
Smardon, A. et al. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr. Biol. 10, 169–178 (2000).
Cogoni, C. & Macino, G. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature 399, 166–169 (1999).
Dalmay, T., Hamilton, A. J., Rudd, S., Angell, S. & Baulcombe, D. C. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101, 543–553 (2000).
Mourrain, P. et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101, 533–542 (2000).
Iyer, L. M., Koonin, E. V. & Aravind, L. Evolutionary connection between the catalytic subunits of DNA-dependent RNA polymerases and eukaryotic RNA-dependent RNA polymerases and the origin of RNA polymerases. BMC Struct. Biol. 3, 1–23 (2003).
Dalmay, T. D., Horsefield, R., Braunstein, T. H. & Baulcombe, D. C. SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. EMBO J. 20, 2069–2078 (2001).
Xie, Z., Fan, B., Chen, C. H. & Chen, Z. An important role of an inducible RNA-dependent RNA polymerase in plant antiviral defense. Proc. Natl Acad. Sci. USA 98, 6516–6521 (2001).
Makeyev, E. V. & Bamford, D. H. Cellular RNA-dependent RNA polymerase involved in posttranscriptional gene silencing has two distinct activity modes. Mol. Cell 10, 1417–1427 (2002).
Schiebel, W., Haas, B., Marinkovic, S., Klanner, A. & Sanger, H. L. RNA-directed RNA polymerase from tomato leaves. II. Catalytic in vitro properties. J. Biol. Chem. 268, 11858–11867 (1993).
Vaistij, F. E., Jones, L. & Baulcombe, D. C. Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell 14, 857–867 (2002).
Voinnet, O., Vain, P., Angell, S. & Baulcombe, D. C. Systemic spread of sequence-specific transgene RNA degradation is initiated by localized introduction of ectopic promoterless DNA. Cell 95, 177–187 (1998).
Yamada, K. et al. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302, 842–846 (2003).
Martienssen, R. Maintenance of heterochromatin by RNA interference of tandem repeats. Nature Genet. 35, 1–2 (2003).
Voinnet, O., Lederer, C. & Baulcombe, D. C. A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103, 157–167 (2000).
Palauqui, J.-C., Elmayan, T., Pollien, J.-M. & Vaucheret, H. Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J. 16, 4738–4745 (1997).
Voinnet, O. & Baulcombe, D. C. Systemic signalling in gene silencing. Nature 389, 553 (1997).
Timmons, L. & Fire, A. Specific interference by ingested dsRNA. Nature 395, 854 (1998).
Feinberg, E. H. & Hunter, C. P. Transport of dsRNA into cells by the transmembrane protein SID-1. Science 301, 1545–1547 (2003).
Winston, W. M., Molodowitch, C. & Hunter, C. P. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science 295, 2456–2459 (2002).
Haywood, V., Kragler, F. & Lucas, W. J. Plasmodesmata: pathways for protein and ribonucleoprotein signaling. Plant Cell 14, S303–S325 (2002).
Himber, C., Dunoyer, P., Moissiard, G., Ritzenthaler, C. & Voinnet, O. Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J. 22, 4523–4533 (2003).
Lakatos, L., Szittya, G., Silhavy, D. & Burgyan, J. Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses. EMBO J. 23, 876–884 (2004).
Mallory, A. C. et al. HC-Pro suppression of transgene silencing eliminates the small RNAs but not transgene methylation or the mobile signal. Plant Cell 13, 571–583 (2001).
Ding, B., Kwon, M.-O., Hammond, R. & Owens, R. Cell-to-cell movement of potato spindle tuber viroid. Plant J. 12, 931–936 (1997).
Kim, M., Canio, W., Kessler, S. & Sinha, N. Developmental changes due to long-distance movement of a homeobox fusion transcript in tomato. Science 293, 287–289 (2001).
Juarez, M. T., Kul, J. S., Thomas, J., Heller, B. A. & Timmermans, M. C. microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428, 84–88 (2004).
Kidner, C. A. & Martienssen, R. Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 428, 81–84 (2004).
Yoo, B.-C. et al. A systemic small RNA signaling system in plants. Plant Cell, 1979–2000 (2004).
Moissiard, G. & Voinnet, O. Viral suppression of RNA silencing in plants. Mol. Plant Pathol. 5, 71–82 (2004).
Szittya, G., Molnar, A., Silhavy, D., Hornyik, C. & Burgyan, J. Short defective interfering RNAs of tombusviruses are not targeted but trigger post-transcriptional gene silencing against their helper virus. Plant Cell 14, 359–372 (2002).
Wang, M.-B. et al. On the role of RNA silencing in the pathogenicity and evolution of viroids and viral satellites. Proc. Natl Acad. Sci. USA 101, 3275–3280 (2004).
Chapman, E. J., Prokhnevsky, A. I., Gopinath, K., Dolja, V. & Carrington, J. C. Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev. 18, 1179–1186 (2004).
Dunoyer, P., Lecellier, C. H., Parizotto, E. A., Himber, C. & Voinnet, O. Probing the microRNA and small interfering RNA pathways with virus-encoded suppressors of RNA silencing. Plant Cell 16, 1235–1250 (2004).
Mallory, A. C., Reinhart, B. J., Bartel, D., Vance, V. B. & Bowman, L. H. A viral suppressor of RNA silencing differentially regulates the accumulation of short interfering RNAs and microRNAs in tobacco. Proc. Natl Acad. Sci. USA 99, 15228–15233 (2002).
Vargason, J. M., Szittya, G., Burgyan, J. & Tanaka Hall, T. M. Size selective recognition of siRNA by an RNA silencing suppressor. Cell 115, 799–811 (2003).
Ye, K., Malinina, L. & Patel, D. J. Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature 426, 874–878 (2003).
Li, W.-X. et al. Interferon antagonist proteins of influenza and vaccina virus are suppressors of RNA silencing. Proc. Natl Acad. Sci. USA 101, 1350–1355 (2004).
Schwarz, D. S. et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115, 199–208 (2003).
Li, H., Li, W. X. & Ding, S. W. Induction and suppression of RNA silencing by an animal virus. Science 296, 1319–1321 (2002).
Pfeffer, S. et al. Identification of virus-encoded microRNAs. Science 304, 734–736 (2004).
Lichner, Z., Silhavy, D. & Burgyan, J. Double-stranded RNA-binding proteins could suppress RNA interference-mediated antiviral defences. J. Gen. Virol 84, 975–980 (2003).
Kasschau, K. D. et al. P1/HC-Pro a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell 4, 205–217 (2003).
Pruss, G. J. et al. The potyviral suppressor of RNA silencing confers enhanced resistance to multiple pathogens. Virology 320, 107–120 (2004).
Achard, P., Herr, A. J., Baulcombe, D. & Harberd, N. P. Modulation of photoperiodic control of floral transition by a hormonally regulated microRNA. Genes Dev. (in the press).
Aukerman, M. J. & Sakai, H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15, 2730–2741 (2003).
VanHoudt, H., Ingelbrecht, I., VanMontagu, M. & Depicker, A. Post-transcriptional silencing of a neomycin phosphotransferase II transgene correlates with the accumulation of unproductive RNAs and with increased cytosine methylation of 3′ flanking regions. Plant J. 12, 379–392 (1997).
Acknowledgements
I thank the Gatsby Charitable Foundation for supporting work in my laboratory.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The author declares no competing financial interests.
Rights and permissions
About this article
Cite this article
Baulcombe, D. RNA silencing in plants. Nature 431, 356–363 (2004). https://doi.org/10.1038/nature02874
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature02874