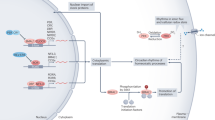
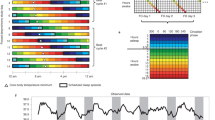
Abstract
Time in the biological sense is measured by cycles that range from milliseconds to years. Circadian rhythms, which measure time on a scale of 24 h, are generated by one of the most ubiquitous and well-studied timing systems. At the core of this timing mechanism is an intricate molecular mechanism that ticks away in many different tissues throughout the body. However, these independent rhythms are tamed by a master clock in the brain, which coordinates tissue-specific rhythms according to light input it receives from the outside world.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Reppert, S. M. & Weaver, D. R. Molecular analysis of mammalian circadian rhythms. Ann. Rev. Physiol. 63, 647–676 (2001)
Bunney, W. E. & Bunney, B. G. Molecular clock genes in man and lower animals: possible implications for circadian abnormalities in depression. Neuropsychopharmacolgy 22, 335–345 (2000)
Welsh, D. K., Logothetis, D. E., Meister, M. & Reppert, S. M. Individual neurons dissociated from rat suprachiasmatic nucleus express independently phase circadian firing rhythms. Neuron 14, 697–706 (1995)
Jagota, A., de la Iglesia, H. O. & Schwartz, W. J. Morning and evening circadian oscillations in the suprachiasmatic nucleus in vitro. Nature Neurosci. 3, 372–376 (2000)
Low-Zeddies, S. S. & Takahashi, J. S. Chimera analysis of the Clock mutation in mice shows that complex cellular integration determines circadian behaviour. Cell 105, 25–42 (2001)
Hamada, T., LeSauter, J., Venuti, J. M. & Silver, R. Expression of Period genes: rhythmic and nonrhythmic compartments of the suprachiasmatic nucleus pacemaker. J. Neurosci. 21, 7742–7750 (2001)
Balsalobre, A., Damiola, F. & Schibler, U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93, 929–937 (1998)
Zylka, M. J., Shearman, L. P., Weaver, D. R. & Reppert, S. M. Three period homologs in mammals: differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron 20, 1103–1110 (1998)
Yamazaki, S. et al. Resetting central and peripheral circadian oscillators in transgenic rats. Science 288, 682–685 (2000)
Pando, M. P., Morse, D., Cermakian N. & Sassone-Corsi, P. Phenotypic rescue of a peripheral clock genetic defect via SCN hierarchical dominance. Cell 110, 107–117 (2002)
Yagita, K., Tamanini, F., van Der Horst, G. T. & Okamura, H. Molecular mechanisms of the biological clock in cultured fibroblasts. Science 292, 278–281 (2001)
Young, M. W. & Kay, S. A. Time zones: a comparative genetics of circadian clocks. Nature Rev. Genet. 2, 702–715 (2001)
Shimomura, K. et al. Genome-wide epistatic interaction analysis reveals complex genetic determinants of circadian behaviour in mice. Genome Res. 11, 959–980 (2001)
King, D. P. et al. Positional cloning of the mouse circadian Clock gene. Cell 89, 641–653 (1997)
Gekakis, N. et al. Role of the CLOCK protein in the mammalian circadian mechanism. Science 280, 1564–1569 (1998)
Hogenesch, J. B., Gu, Y.-Z., Jain, S. & Bradfield, C. A. The basic helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc. Natl Acad. Sci. USA 95, 5474–5479 (1998)
Bunger, M. K. et al. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 103, 1009–1017 (2000)
Kume, K. et al. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98, 193–205 (1999)
Vitaterna, M. H. et al. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc. Natl Acad. Sci. USA 96, 12114–12119 (1999)
Okamura, H. et al. Photic induction of mPer1 and mPer2 in cry-deficient mice lacking a biological clock. Science 286, 2531–2534 (1999)
Shearman, L. P. et al. Interacting molecular loops in the mammalian circadian clock. Science 288, 1013–1019 (2000)
Oishi, K., Fukui, H. & Ishida, N. Rhythmic expression of BMAL1 mRNA is altered in Clock mutant mice: differential regulation in the suprachiasmatic nucleus and peripheral tissues. Biochem. Biophys. Res. Commun. 268, 164–171 (2000)
Preitner, N. et al. The orphan nuclear receptor REV-ERBa controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110, 251–260 (2002)
Ueda, H. R. et al. A transcription factor response element for gene expression during circadian night. Nature 418, 534–539 (2002)
Yu, W., Nomura, M. & Ikea, M. Interacting feedback loops within the mammalian clock: BMAL1 is negatively autoregulated and upregulated by CRY1, CRY2, and PER2. Biochem. Biophys. Res. Comm. 290, 933–942 (2002)
Bae, K. et al. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron 30, 525–536 (2001)
Zheng, B. et al. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell 105, 683–694 (2001)
van der Horst, G. T. J. et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 398, 627–630 (1999)
Zheng, B. et al. The mPer2 gene encodes a function component of the mammalian clock. Nature 400, 169–173 (1999)
Cermakian, N., Monaco, L., Pando, M. P., Dierich, A. & Sassone-Corsi, P. Altered behavioural rhythms and clock gene expression in mice with a targeted mutation in the Period1 gene. EMBO J. 20, 3967–3974 (2001)
Shearman, L. P., Jin, X., Lee, C., Reppert, S. M. & Weaver, D. R. Targeted disruption of the mPer3 gene: Subtle effects on circadian clock function. Mol. Cell. Biol. 20, 6269–6275 (2000)
Lee, C., Etchegaray, J. P., Cagampang, F. R., Loudon, A. S. & Reppert, S. M. Posttranslational mechanisms regulate the mammalian circadian clock. Cell 107, 855–867 (2001)
Eide, E. J., Vielhaber, E. L., Hinz, W. A. & Virshup, D. M. The circadian regulatory protein BMAL1 and cryptochromes are substrates of casein kinase 1ɛ (CKIɛ). J. Biol. Chem. 277, 17248–17254 (2002)
Sanada, K., Okano, T. & Fukada, Y. Mitogen-activated protein kinase phosphorylates and negatively regulates basic helix-loop-helix-PAS transcription factor BMAL1. J. Biol. Chem. 277, 267–271 (2002)
Lowrey, P. L. et al. Positional syntenic cloning and functional characterization of a mammalian circadian mutation tau. Science 288, 483–491 (2000)
Toh, K. L. et al. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science 291, 1040–1043 (2001)
Akashi, M., Tsuchiya, Y., Yoshino, T. & Nishida, E. Control of intracellular dynamics of mammalian period proteins by casein kinase I epsilon (CKIɛ) and CKIδ in cultured cells. Mol. Cell. Biol. 22, 1693–1703 (2002)
Martinek, S., Inonog, S., Manoukian, A. S. & Young, M. W. A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell 105, 769–779 (2001)
Vielhaber, E. L., Duricka, D., Ullman, K. S. & Virshup, D. M. Nuclear export of mammalian PERIOD proteins. J. Biol. Chem. 276, 45921–45927 (2001)
Yagita, K. et al. Nucleocytoplasmic shuttling and mCRY-dependent inhibition of ubiquitylation of the mPER2 protein. EMBO J. 21, 1301–1314 (2002)
McNamara, P., Seo, S.-B., Rudic, R. D., Sehgal, A., Chakravarti, D. & FitzGerald, G. A. Regulation of CLOCK and MOP4 by nuclear hormone receptors in the vasculature: a humoral mechanism to reset a peripheral clock. Cell 105, 877–889 (2001)
Rutter, J., Reick, M., Wu, L. C. W. & McKnight, S. L Regulation of CLOCK and NPAS2 DNA binding by the redox state of NAD cofactors. Science 293, 510–514 (2001)
Zhang, Q., Piston, D. W. & Goodman, R. H. Regulation of corepressor function by nuclear NADH. Science 295, 1895–1897 (2002)
Wright, K. P. Jr & Czeisler, C. A. Absence of circadian phase resetting in response to bright light behind the knee. Science 297, 571 (2002)
Lucas, R. J. et al. Identifying the photoreceptive inputs to the mammalian circadian system using transgenic and retinally degenerate mice. Behav. Brain Res. 125, 97–102 (2001)
Provencio, I. et al. A novel human opsin in the inner retina. J. Neurosci. 20, 600–605 (2000)
Hattar, S., Liao, H. W., Takao, M., Berson, D. M. & Yau, K. W. Melanopsin-containing retinal ganglion cells: architecture, projections, and intrinsic photosensitivity. Science 295, 1065–1070 (2002)
Provencio, I., Rollag, M. D. & Castrucci, A. M. Photoreceptive net in the mammalian retina. Nature 415, 493 (2002)
Gooley, J. J., Lu, J., Chou, T. C., Scammell, T. E. & Saper, C. B. Melanopsin in cells of origin of the retinohypothalamic tract. Nature Neurosci. 4, 1165 (2001)
Hannibal, J., Hindersson, P., Knudsen, S. M., Georg, B. & Fahrenkrug, J. The photopigment melanopsin is exclusively present in pituitary adenylate cyclase-activating polypeptide-containing retinal ganglion cells of the retinohypothalamic tract. J. Neurosci. 22, RC191 (2002)
Berson, D. M., Dunn, F. A. & Takao, M. Phototransduction by retinal ganglion cells that set the circadian clock. Science 295, 1070–1073 (2002)
Selby, C. P., Thompson, C., Schmitz, T. M., Van Gelder, R. N. & Sancar, A. Functional redundancy of cryptochromes and classical photoreceptors for nonvisual ocular photoreception in mice. Proc. Natl Acad. Sci. USA 97, 14697–14702 (2000)
Thompson, C. L. et al. Preservation of light signaling to the suprachiasmatic nucleus in vitamin A-deficient mice. Proc. Natl Acad. Sci. USA 98, 11708–11713 (2001)
Griffin, E. A., Staknis, D. & Weitz, C. J. Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science 286, 768–771 (1999)
Shigeyoshi, Y. et al. Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript. Cell 91, 1043–1053 (1997)
Albrecht, U., Zheng, B., Larkin, D., Sun, Z. S. & Lee, C. C. mPer1 and mPer2 are essential for normal resetting of the circadian clock. J. Biol. Rhythms 16, 100–104 (2001)
Field, M. D. et al. Analysis of clock proteins in mouse SCN demonstrates phylogenetic divergence of the circadian clockwork and resetting mechanisms. Neuron 251, 437–447 (2000)
Crosio, C., Cermakian, N., Allis, C. D. & Sassone-Corsi, P. Light induced chromatin modification in cells of the mammalian circadian clock. Nature Neurosci. 3, 1241–1247 (2000)
Travnickova-Bendova, Z., Cermakian, N., Reppert, S. M. & Sassone-Corsi, P. Bimodal regulation of mPeriod promoters by CREB-dependent signaling and CLOCK:BMAL1 activity. Proc. Natl Acad. Sci. USA 99, 7728–7733 (2002)
Shearman, L. P. & Weaver, D. R. Photic induction of Period gene expression is reduced in Clock mutant mice. NeuroReport 10, 613–618 (1999)
Gau, D. et al. Phosphorylation of CREB Ser-142 regulates light-induced phase shifts of the circadian clock. Neuron 34, 245–253 (2002)
Pennartz, C. M. A., de Jeu, M. T. G., Bos, N. P. A., Schaap, J. & Geurtsen, A. M. S. Diurnal modulation of pacemaker potentials and calcium current in the mammalian circadian clock. Nature 416, 286–290 (2002)
Kramer, A. et al. Regulation of daily locomotor activity and sleep by hypothalamic EGF receptor signaling. Science 294, 2511–2515 (2001)
Cheng, M. Y. et al. Prokineticin 2 transmits the behavioural circadian rhythm of the suprachiasmatic nucleus. Nature 417, 405–410 (2002)
Bartness, T. J., Song, C. K. & Demas, G. E. SCN efferents to peripheral tissues: implications for biological rhythms. J. Biol. Rhythms 16, 196–204 (2001)
Balsalobre, A. et al. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science 289, 2344–2347 (2000)
Damiola, F. et al. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 14, 2950–2961 (2000)
Stokkan, K. A., Yamazaki, S., Tei, H., Sakaki, Y. & Menaker, M. Entrainment of the circadian clock in the liver by feeding. Science 291, 490–493 (2001)
Le Minh, N., Damiloa, F., Tronche, F., Schutz, G. & Schibler, U. Glucocorticoids inhibit food-induced phase-shifting in peripheral circadian oscillators. EMBO J. 20, 7128–7136 (2001)
Jin, X. et al. A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell 96, 57–68 (1999)
Ripperger, J. A. et al. CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP. Genes Dev. 14, 679–689 (2000)
Lavery, D. J. et al. Circadian expression of the steroid 15 α-hydroxylase (Cyp2a4) and coumarin 7-hydroxylase (Cyp2a5) genes in mouse liver is regulated by the PAR leucine zipper transcription factor DBP. Mol. Cell. Biol. 19, 6488–6499 (1999)
Mitsui, S., Yamaguchi, S., Matsuo, T., Ishida, Y. & Okamura, H. Antagonistic role of E4BP4 and PAR proteins in the circadian oscillatory mechanism. Genes Dev. 15, 995–1006 (2001)
Clayton, J. D., Kyriacou, C. P. & Reppert, S. M. Keeping time with the human genome. Nature 409, 829–831 (2000)
Grundschober, C. et al. Circadian regulation of diverse gene products revealed by mRNA expression profiling of synchronized fibroblasts. J. Biol. Chem. 276, 46751–46758 (2001)
Duffield, G. E. et al. Circadian programs of transcriptional activation, signaling, and protein turnover revealed by microarray analysis of mammalian cells. Curr. Biol. 12, 551–557 (2002)
Kita, Y. et al. Implications of circadian gene expression in kidney, liver and the effects of fasting on pharmacogenomic studies. Pharmacogenetics 12, 55–65 (2002)
Akhtar, R. A. et al. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr. Biol. 12, 540–550 (2001)
Panda, S. et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109, 307–320 (2002)
Storch, K. F. et al. Extensive and divergent circadian gene expression in liver and heart. Nature 417, 78–83 (2002)
Harmar, A. J. et al. The VPAC2 receptor is essential for circadian function in the mouse suprachiasmatic nuclei. Cell 109, 497–508 (2002)
Reick, M., Garcia, J. A., Dudley, C. & McKnight, S. L. NPAS2: an analog of clock operative in the mammalian forebrain. Science 293, 506–509 (2001)
Liu, C. & Reppert, S. M. GABA synchronizes clock cells within the suprachiasmatic circadian clock. Neuron 25, 123–128 (2000)
Acknowledgements
We thank members of the Reppert laboratory for contributing to the ideas presented in this review, and U. Schibler for sharing data before publication. This work was supported by grants from the NIH and DARPA.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Reppert, S., Weaver, D. Coordination of circadian timing in mammals. Nature 418, 935–941 (2002). https://doi.org/10.1038/nature00965
Issue Date:
DOI: https://doi.org/10.1038/nature00965