Abstract
Spinocerebellar ataxias (SCAs) are a heterogeneous group of disorders with almost 30 subtypes. The prevalence and relative frequency of each subtype vary among different populations. In this article, we report the relative frequency of six SCA subtypes in the Thai population and attempt to explain the observed pattern when compared with other populations in this region. We searched for SCA type 1, SCA2, SCA3, SCA6, SCA7 and dentatorubral–pallidoluysian atrophy mutations using GeneScan analysis in 340 patients from 182 families, in which at least one person had a clinical diagnosis of SCA. We analyzed the relative frequencies of SCA subtypes on a family basis, and compared these with the data in the Chinese and Indian populations. SCA3 was found in 19.2% of the patients (Agresti–Coull 95% confidence interval: 14.1–25.6%), SCA1 in 11.5% (7.6–17.1%) and SCA2 in 10.4% (6.7–15.8%). SCA6 was found in three families, with a relative frequency of 1.6% (0.3–5.0%). Compared with the related populations, the Thai SCA3 frequency was less than that of the Chinese, whereas it was higher than that in most of the Indian studies. The reverse is true for the SCA1/SCA2 frequency. A similar study in Singapore, where there was a clear history of population admixture, also showed the frequencies between those of the Chinese and the Indian populations. Although SCA3 was the most common identifiable SCA subtype in Thailand, SCA1 and SCA2 were also relatively common. Our results also supported some degree of admixture with the Indians in the Thai population and justify further study in the area.
Similar content being viewed by others
Introduction
Spinocerebellar ataxias (SCAs), also known clinically as autosomal dominant cerebellar ataxias, are a group of genetically and phenotypically heterogeneous ataxic disorders resulting from the progressive degeneration of the cerebellum and its connections, with an estimated prevalence of up to five to seven cases per 100 000 people.1, 2, 3 The most widely accepted classification of SCAs at present corresponds to the order of gene description1 and consists of at least 28 subtypes, corresponding to defects in at least 28 distinct loci.2 Among these, SCA3 (also known as Machado–Joseph disease: MJD) is the most common subtype worldwide, whereas SCA1, SCA2, SCA6, SCA7 and SCA8 are the other common subtypes, each with over 2% prevalence; the rest are believed to be rare.3
The overall prevalence and frequency of genetic subtypes of SCA vary among different populations, similar to the case in Cuba (predominantly SCA2), Portugal, Brazil and Azores (SCA3/MJD), Mexico (SCA10) and Japan (dentatorubral–pallidoluysian atrophy: DRPLA).1, 2, 3 Estimations of the subtype frequencies are therefore important in the planning of diagnostic strategy in a specific population. Furthermore, as the frequency of each SCA subtype also differs between the Chinese-related and Indian-related populations (see Discussion), and the variation is believed to be the result of founder populations,3, 4 it could potentially be useful for assessing the mix between the Chinese and the Indians in the Indochina peninsula.
Materials and methods
Study design
This is a retrospective review of the results collected as part of genetic consultation from August 1994 to July 2006. The study has been approved by the ethical committee of Ramathibodi Hospital.
Subjects
Our medical genetic division has offered molecular diagnostic service for SCAs since 1994. By July 2006, we had obtained DNA samples of 340 individuals from 182 families. Each family has at least one person who had symptoms of progressive ataxia unexplained by other metabolic, nutritional or infective causes, and without space-occupying lesion on the cranial magnetic resonance imaging. These patients were clinically diagnosed as having SCAs by their attending neurologists and were referred to the genetic service for molecular confirmation and for counseling about the disease and the risks to themselves and their relatives. As part of the counseling, patients were advised to share the information with their relatives, who can then opt for a counseling session with a view to undergo predictive testing if they so wish. This is the reason why some of the non-index cases were asymptomatic at the time of testing. If more than one symptomatic person in a family attended the service at the same time, we assigned only one person as the index case and the remaining as non-index cases (see also Discussion).
DNA analysis
Genomic DNA of each subject was extracted from the peripheral blood sample by the standard phenol–chloroform method. PCR was performed with the primers Rep1 and Rep2 for SCA15 (annealing temperature, 62 °C), SCA2-A and SCA2-B for SCA26 (57 °C), MJD52 and MJD25 for SCA3/MJD7 (57 °C), SCA6F and SCA6R for SCA68 (57 °C), 4U1024 and 4U716 for SCA79 (52 °C) and CTG-B37-F and CTG-B37-R for DRPLA10, 11 (47 °C). The 5′-end of each forward primer was fluorescent-tagged, allowing us to determine the repeat size of each amplicon by GeneScan analysis (ABI 310 Genetic analyzer; Applied Biosystems, Foster City, CA, USA). Subjects with more than 40 CAG repeats in SCA1, 34 repeats in SCA2, 55 repeats in SCA3/MJD, 21 repeats in SCA6, 37 repeats in SCA7 or 48 CAG repeats in DRPLA locus were classified as having positive results for that subtype.
Between 1994 and 2004, only diagnostic testing for SCA1 was offered. When we introduced tests for SCA3/MJD (2004) and SCA6 (late 2005), the stored DNA samples of the patients who had negative test result for SCA1 were reanalyzed for SCA3/MJD and SCA6. More recently, the remaining negative samples were again retested when the tests for SCA2, SCA7 and DRPLA became available.
Data entry and analysis
The test results, along with the individual and family identification data, were entered into a spreadsheet application. Validation and cleaning was carried out before importation into R-2.8.012 for statistical analysis. After initial data integrity check, we calculated the proportion of each SCA subtype with its Agresti–Coull 95% confidence interval13 in all subjects as well as in only the index cases, and compared these with those of earlier studies in related populations. We also calculated the ratio between the risk of having SCA3 and that of having either SCA1 or SCA2 (that is, the odds of having SCA3 rather than SCA1/2) in each population, along with its non-parametric bootstrap bias-corrected and accelerated (BCa) 95% confidence interval, and compared this across the populations.
Results
Among the 340 individuals tested, 182 were index cases in their families and 158 were the relatives of the index cases. They came from 182 families, with the numbers of individuals tested in each family ranging from 1 to 20. All of the subjects were Thai; the majority of them were from the Bangkok area (see Table 1). As we have relatively few subjects outside the Bangkok area, stratification analysis by region was not very informative. The relative frequency among the Bangkok cases was similar to the overall frequency.
The frequency of each SCA subtype is shown in Table 1. The most prevalent subtype detected was SCA3/MJD, which affected 19% of the families, followed by SCA1 (11.5%) and SCA2 (10.4%). SCA6 was relatively rare, with just less than 2% in relative frequency. About 57% of the index cases could not be assigned a molecular diagnosis based on the tests currently offered, implying other disease subtypes.
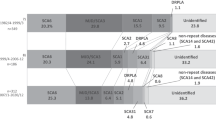
Comparison of the subtype frequencies with other populations is shown in Table 2 and graphically in Figure 1.
Geographical variation in SCA subtypes in south, southeast and east Asia (a), and the odds of having the ‘Chinese SCA’ (SCA3) rather than ‘Indian SCAs’ (SCA1 and -2) cases in these populations, with their non-parametric bootstrap BCa 95% confidence intervals (b). The Hong Kong study was excluded from the geographical plot as the number of negative (‘other’) cases had not been reported. The upper confidence limits were 10 and 12 for China and Hong Kong, respectively. The confidence interval for the East India/Bihar study could not be calculated due to the total absence of SCA3 case within the sample. See Table 2 for data sources.
Discussion
In line with earlier studies in most populations, we found that SCA3/MJD is the most prevalent subtype of SCAs in the Thai population. However, this accounted for just about 19% of the families compared with about 30% in the Chinese-related populations other than those in Singapore. On the contrary, apart from the studies carried out in West Bengal, which reported the relative frequency of SCA3/MJD of approximately 36%,24 the reported relative frequency of SCA3/MJD ranged from 0 to about 14% in most studies in Indian populations.19, 20, 21, 23 The West Bengal study is in fact an interesting exception, as earlier studies have shown a genetic relationship between the population in this area and the Tibeto-Burman speakers from Southeast and East Asia.26, 27, 28, 29 Also, interestingly, in Singaporeans who a are mixture of ethnic Chinese, Malays and Indians, the relative frequency of SCA3/MJD was even lower than that in ours, at about 13%.
At 11.5%, the relative frequency of SCA1 in our population was near the lower end of the relative frequencies in the Indians (7–32%),19, 20, 21, 22, 23 but was much higher than those in the Chinese/Japanese populations (approximately 3%).15, 17, 18, 25, 30 SCA2, which was almost as common as SCA1 in our population, also showed a similar pattern: its relative frequency of 10.4% was much lower than that in the Indians (18–57%)19, 20, 21, 22, 23 while being still marginally higher than the upper range of the frequencies in the Chinese populations (4–10%).15, 17, 18, 25, 30
These three common subtypes accounted for 41% of the cases. Of the remaining three tested subtypes, only SCA6 was found. SCA6 is generally rare in this region, apart from in a specific ethnic group in Taiwan. Our relative frequency of SCA6 was comparable with that of the Singaporeans.
The frequency of index patients who were not detected as SCA1, -2, -3, -6, -7 or DRPLA in this study was comparable with that from other studies in the region. This probably reflected the prevalence of one or more untested subtypes such as SCA12.23, 25
The number of all individuals who had negative results could be partly inflated as asymptomatic (thus possibly unaffected) family members were included. As the frequency of SCA subtypes in all cases would also be influenced by the size of the families, we believe the analysis based on the index cases as representatives for their families as carried out here would yield a more accurate result.
An interesting pattern emerges if one compares, among populations in this region, the odds of having SCA3, which is the predominant type in Chinese populations, rather than having either SCA1 or SCA2, which are more common among the Indian populations. In our series, the odds were 0.88, which was similar to those in the other two populations with evident admixture (Singapore25 and West Bengal;24 see Figure 1b). With a range of 0.83 to 0.94, the odds in these three populations were much higher than those in the Indian populations (highest value=0.28 in South India 22), whereas being much lower than those in the Chinese populations (lowest value=2.60 in Taiwan18). We believe that this pattern could support the hypothesis that there is an admixture between the Chinese- and Indian-related populations in the Thai population. However, some caution is needed against making conclusions based on this alone, as many studies had small sample sizes, making the estimations rather unstable. Furthermore, although this pattern will arise in the presence of a population admixture, the reverse is not necessarily true: it could arise, for example, if the Thai SCAs have a background independent of the Indian or Chinese populations, or because of a random founding population within the main population, or even a random error from sampling.
Over the last decade, genetic studies have linked the populations in the Indochina peninsula, including the Thai people, to the Han Chinese, particularly those in the southern part of China.31, 32, 33 This is quite expected as the region is close to China and there has earlier been historical, archeological and linguistic evidence to support the relationship.34, 35 At the same time, given the similar proximity of the region to India, and the historical, archeological and cultural evidence of relationship with the Indian subcontinent,34, 36 it is very likely that there is some degree of mixing between the Chinese and Indian populations in the Indochina peninsula, although there has earlier been little genetic evidence to support this. In this context, we believe that the observation from this study at least indirectly adds genetic support for this historical and cultural linking.
The majority of our subjects came from Bangkok and the central part of Thailand. Although this could mean that people in this area are relatively overrepresented in our sample, the degree of overrepresentation may not be as high, as the official population data are known to underestimate the number of people in the urban area, particularly Bangkok and its surroundings, and overestimate the number of people in a more rural area. This is because of the ‘unofficial’ migration that has not been accounted for in the official registry. Furthermore, because of the relatively small size of the country and the homogeneity of the population, we do not think that the true relative frequencies in the Thai population would differ much from our estimation.
On the basis of the subtype frequencies from this study, it would be prudent to look for SCA3 first, followed by SCA1 and -2 in a Thai patient with presumed diagnosis of SCA unless the clinical features suggest otherwise, or when a “panel” test is used. In the latter case, these three common types should be included in the panel as a minimum. As for the population inference, it is not possible to infer the direction of migration or the timing from this study. Nevertheless, we believe this provides enough justification for further, more definitive molecular studies to look into the link between the populations on the Indochina peninsula and the Indians.
References
Manto, M.-U. The wide spectrum of spinocerebellar ataxias (SCAs). Cerebellum 4, 2–6 (2005).
Dueñas, A. M., Goold, R. & Giunti, P. Molecular pathogenesis of spinocerebellar ataxias. Brain 129, 1357–1370 (2006).
Schöls, L., Bauer, P., Schmidt, T., Schulte, T. & Riess, O. Autosomal dominant cerebellar ataxias: clinical features, genetics, and pathogenesis. Lancet Neurol. 3, 291–304 (2004).
Martins, S., Calafell, F., Gaspar, C., Wong, V. C., Silveira, I., Nicholson, G. A. et al. Asian origin for the worldwide-spread mutational event in Machado–Joseph disease. Arch. Neurol. 64, 1502–1508 (2007).
Orr, H. T., Chung, M. Y., Banfi, S., Kwiatkowski, T. J. Jr, Servadio, A., Beaudet, A. L. et al. Expansion of an unstable trinucleotide CAG repeat in spinocerebellar ataxia type I. Nat. Genet. 4, 221–226 (1993).
Pulst, S. M., Nechiporuk, A., Nechiporuk, T., Gispert, S., Chen, X. N., Lopes-Cendes, I. et al. Moderate expansion of a normally biallelic trinucleotide repeat in spinocerebellar ataxia type 2. Nat. Genet. 14, 269–276 (1996).
Kawaguchi, Y., Okamoto, T., Taniwaki, M., Aizawa, M., Inoue, M., Katayama, S. et al. CAG expansions in a novel gene for Machado–Joseph disease at chromosome 14q32.1. Nat. Genet. 8, 221–227 (1994).
Michlewski, G. & Krzyzosiak, W. J. Molecular architecture of CAG repeats in human disease related transcripts. J. Mol. Biol. 340, 665–679 (2004).
David, G., Abbas, N., Stevanin, G., Dürr, A., Yvert, G., Cancel, G. et al. 1997. Cloning of the SCA7 gene reveals a highly unstable CAG repeat expansion. Nat. Genet. 17, 65–70 (1997).
Li, S.-H., McInnis, M. G., Margolis, R. L., Antonarakis, S. E. & Ross, C. A. Novel triplet repeat containing genes in human brain: cloning, expression, and length polymorphisms. Genomics 16, 572–579 (1993).
Koide, R., Ikeuchi, T., Onodera, O., Tanaka, H., Igarashi, S., Endo, K. et al. Unstable expansion of CAG repeat in hereditary dentatorubral–pallidoluysian atrophy (DRPLA). Nat. Genet. 6, 9–13 (1994).
R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing (2008) (Available online at URL: http://www.R-project.org).
Agresti, A. & Coull, B. A. Approximate is better than ‘exact’ for interval estimation of binomial proportions. Am. Stat. 52, 119–126 (1998).
Bureau of Registry Administration, Department of Provincial Administration. (Notification of the Bureau of Central Registry, Department of Provincial Administration, on the number of citizens throughout the Kingdom divided into the Bangkok Metropolis and each other province, according to the Citizen Registry documentation as of 31st December, B. E. 2550 (2007). Royal Gazette 125 (suppl 25 Ngor), 7–10 (2008) (in Thai).
Tang, B., Liu, C., Shen, L., Dai, H., Pan, Q., Jing, L. et al. Frequency of SCA1, SCA2, SCA3/MJD., SCA6, SCA7, and DRPLA CAG trinucleotide repeat expansion in patients with hereditary spinocerebellar ataxia from Chinese kindreds. Arch. Neurol. 57, 540–544 (2000).
Lau, K. K., Lam, K., Shiu, K. L., Au, K. M., Tsoi, T. H., Chan, A. Y. W. et al. Clinical features of hereditary spinocerebellar ataxia diagnosed by molecular genetic analysis. Hong Kong Med. J. 10, 255–259 (2004).
Soong, B. W., Lu, Y. C., Choo, K. B. & Lee, H. Y. Frequency analysis of autosomal dominant cerebellar ataxias in Taiwanese patients and molecular characterization of spinocerebellar ataxia type 6. Arch. Neurol. 58, 1105–1109 (2001).
Tsai, H.-F., Liu, C.-S., Leu, T.-M., Wen, F.-C., Lin, S.-J., Liu, C.-C. et al. Analysis of trinucleotide repeats in different SCA loci in spinocerebellar ataxia patients and in normal population of Taiwan. Acta Neurol. Scand. 109, 355–360 (2004).
Saleem, Q., Choudhry, S., Mukerji, M., Bashyam, L., Padma, M. V., Chakravarthy, A. et al. Molecular analysis of autosomal dominant hereditary ataxias in the Indian population: high frequency of SCA2 and evidence for a common founder mutation. Hum. Genet. 106, 179–187 (2000).
Basu, P., Chattopadhyay, B., Gangopadhaya, P. K., Mukherjee, S. C., Sinha, K. K., Das, S. K. et al. Analysis of CAG repeats in SCA1, SCA2, SCA3, SCA6, SCA7 and DRPLA loci in spinocerebellar ataxia patients and distribution of CAG repeats at the SCA1, SCA2 and SCA6 loci in nine ethnic populations of eastern India. Hum. Genet. 106, 597–604 (2000).
Sinha, K. K., Worth, P. F., Jha, D. K., Sinha, S., Stinton, V. J., Davis, M. B. et al. Autosomal dominant cerebellar ataxia: SCA2 is the most frequent mutation in eastern India. J. Neurol. Neurosurg. Psychiatr. 75, 448–452 (2004).
Krishna, N., Mohan, S., Yashavantha, B. S., Rammurthy, A., Kiran Kumar, H. B., Mittal, U. et al. SCA 1, SCA 2 & SCA 3/MJD mutations in ataxia syndromes in southern India. Indian J. Med. Res. 126, 465–470 (2007).
Srivastava, A. K., Choudhry, S., Gopinath, M. S., Roy, S., Tripathi, M., Brahmachari, S. K. et al. Molecular and clinical correlation in five Indian families with spinocerebellar ataxia 12. Ann. Neurol. 50, 796–800 (2001).
Chakravarty, A. & Mukherjee, S. C. Autosomal dominant cerebellar ataxias in ethnic Bengalees in West Bengal—an Eastern Indian state. Acta Neurol. Scand. 105, 202–208 (2002).
Zhao, Y., Tan, E. K., Law, H. Y., Yoon, C. S., Wong, M. C. & Ng, I. Prevalence and ethnic differences of autosomal-dominant cerebellar ataxia in Singapore. Clin. Genet. 62, 478–481 (2002).
Su, B., Xiao, C., Deka, R., Seielstad, M. T., Kangwanpong, D., Xiao, J. et al. Y chromosome haplotypes reveal prehistorical migrations to the Himalayas. Hum. Genet. 107, 582–590 (2000).
Basu, A., Mukherjee, N., Roy, S., Sengupta, S., Banerjee, S., Chakraborty, M. et al. Ethnic India: a genomic view, with special reference to peopling and structure. Genome Res. 13, 2277–2290 (2003).
Cordaux, R., Weiss, G., Saha, N. & Stoneking, M. The northeast Indian passageway: a barrier or corridor for human migrations? Mol. Biol. Evol. 21, 1525–1533 (2004).
Kashyap, V. K., Guha, S., Sitalaximi, T., Bindu, G. H., Hasnain, S. E. & Trivedi, R. Genetic structure of Indian populations based on fifteen autosomal microsatellite loci. BMC Genet. 7, 28 (2006).
Takano, H., Cancel, G., Ikeuchi, T., Lorenzetti, D., Mawad, R., Stevanin, G. et al. Close associations between prevalences of dominantly inherited spinocerebellar ataxias with CAG-repeat expansions and frequencies of large normal CAG alleles in Japanese and Caucasian populations. Am. J. Hum. Genet. 63, 1060–1066 (1998).
Chu, J. Y., Huang, W., Kuang, S. Q., Wang, J. M., Xu, J. J., Chu, Z. T. et al. Genetic relationship of populations in China. Proc. Natl Acad. Sci. USA 95, 11763–11768 (1998).
Tajima, A., Pang, I.-H., Fucharoen, G., Fucharoen, S., Matsuo, M., Tokunaga, K. et al. Three major lineages of Asian Y chromosomes: implications for the peopling of east and southeast Asia. Hum. Genet. 110, 80–88 (2002).
Hill, C., Soares, P., Mormina, M., Macaulay, V., Meehan, W., Blackburn, J. et al. Phylogeography and ethnogenesis of aboriginal Southeast Asians. Mol. Biol. Evol. 23, 2480–2491 (2006).
Wyatt, D. K. Thailand: A Short History (Trasvin Publications, Chiang Mai, 1984).
Bellwood, P. The origins and dispersals of agricultural communities in Southeast Asia. In Southeast Asia: From Prehistory to History (eds Glover, I. & Bellwood, P.) 21–40 (RoutledgeCurzon, New York, 2004).
Bellina, B. & Glover, I. The archaeology of early contact with India and the Mediterranean world, from the fourth century BC to the fourth century AD. In Southeast Asia: From Prehistory to History (eds Glover, I. & Bellwood, P.) 68–88 (RoutledgeCurzon, New York, 2004).
Acknowledgements
This study was supported by the annual research grant of the Faculty of Medicine Ramathibodi Hospital, Mahidol University, Thailand. The map in Figure 1a was created using the R packages MAPS 2.0–40 and MAPDATA 2.0–23.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sura, T., Eu-ahsunthornwattana, J., Youngcharoen, S. et al. Frequencies of spinocerebellar ataxia subtypes in Thailand: window to the population history?. J Hum Genet 54, 284–288 (2009). https://doi.org/10.1038/jhg.2009.27
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2009.27