Abstract
Hereditary spastic paraplegias (HSP) are a genetically heterogeneous group of disorders characterized by a distal axonopathy of the corticospinal tract motor neurons leading to progressive lower limb spasticity and weakness. Intracellular membrane trafficking, mitochondrial dysfunction and myelin formation are key functions involved in HSP pathogenesis. Only recently defects in metabolism of complex lipids have been implicated in a number of HSP subtypes. Mutations in the 23 known autosomal recessive HSP genes explain less than half of autosomal recessive HSP cases. To identify novel autosomal recessive HSP disease genes, exome sequencing was performed in 79 index cases with autosomal recessive forms of HSP. Resulting variants were filtered and intersected between families to allow identification of new disease genes. We identified two deleterious mutations in the phospholipase DDHD2 gene in two families with complicated HSP. The phenotype is characterized by early onset of spastic paraplegia, mental retardation, short stature and dysgenesis of the corpus callosum. Phospholipase DDHD2 is involved in intracellular membrane trafficking at the golgi/ endoplasmic reticulum interface and has been shown to possess phospholipase A1 activity in vitro. Discovery of DDHD2 mutations in HSP might therefore provide a link between two key pathogenic themes in HSP: membrane trafficking and lipid metabolism.
Similar content being viewed by others
INTRODUCTION
The genetically diverse group of hereditary spastic paraplegias (HSP) is clinically defined by a progressive spasticity and weakness of the lower limbs, caused by distal axonopathy of the long motor axons of the corticospinal tract. Identification of more than 39 HSP1 genes highlights intracellular membrane trafficking, mitochondrial metabolism and myelin formation as key functions involved in HSP pathogenesis.2 Only recently lipid metabolism has emerged as another main theme in HSP pathophysiology, backed by the discovery of mutations in a number of genes involved in lipid metabolism and signaling including CYP2U1,3 CYP7B1,4 DDHD1,3 FA2H,5 GBA26 and PNPLA6.7
In this study, we have identified deleterious mutations in phospholipase DDHD2, the sister enzyme of PA-PLA1/DDHD1 in two families with complicated HSP.
Materials and METHODS
Exome sequencing was performed in 79 index patients with familial forms of autosomal recessive HSP. The SureSelect Human All Exon 50 Mb kit (Agilent, Santa Clara, CA, USA) was used for in-solution enrichment; exome sequencing was performed using the Hiseq2000 instrument (Illumina, San Diego, CA, USA). Paired-end reads of 100 bp length were produced. BWA and GATK software packages8, 9 were used to align sequence reads to the reference and call variant positions. The data were then imported into Gem.app, a web-based database and analysis tool for next generation sequencing data developed by the group of S Züchner (https://secureforms.med.miami.edu/hihg/gem-app/)10 for further analysis. An average of 82 613 347 sequence reads was produced per sample, 98.7% of which could be aligned to the targeted sequence. Mean coverage was 75.9-fold; 85.5% of the targeted sequence was covered by at least 10 reads. Variants were filtered for impact on the coding sequence, presence of either one homozygous or two heterozygous variants in one gene, frequency in public databases (minor allele frequency in dbSNP135 and NHLBI ESP6500<0.5%), conservation (GERP score>2 or PhastCons score>0.6) and genotyping quality (GATK quality index>30 and genotype quality GQ>30). Additionally, variants segregating in more than two families in Gem.app were removed. In addition to 79 families with complex HSP, Gem.app contained ∼130 families with pure HSP and ∼450 families with non-HSP phenotypes at the time of analysis.
Informed consent was obtained from all individuals and the Institutional Review Boards at the participating medical centers approved the study.
RESULTS
Gene identification
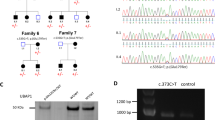
In family THI26003, seven homozygous SNVs and one homozygous Indel were present; two of the SNVs as well as the Indel didn’t segregate with the disease. Segregating homozygous missense variants were found in C14orf166 (NM_016039.2: c.311C>T, p.Pro104Leu; rs149288575), FUT10 (NM_032664.3: c.473T>C, p.Leu158Pro), IMPA1 (NM_001144879.1: c.542C>T, p.Thr181Ile) and OTOGL (NM_173591.3: c.3461A>G, p.Asp1154Gly); a homozygous nonsense variant in DDHD2 (NM_001164234: c.859C>T, p.Arg287*) was the only truncating change present. Details on all the five segregating variants in family THI26003 are given in Supplementary Table 1. As it was not possible to further narrow down the list of candidate genes in family THI26003 alone based on the available in silico parameters, the resulting list of five candidate genes was used as a seed and intersected with the resulting candidate gene lists obtained by exome sequencing in the remaining 78 autosomal recessive HSP families. Another deleterious DDHD2 variant was identified in family IHG25194 (NM_001164232.1: c.1982_1983delAT, p.Tyr661Cysfs*8) that segregates with the disease (Figure 1). Parents of the two affected siblings of this family were not available for genetic analysis, therefore a genomic deletion of one DDHD2 allele cannot be ruled out with certainty. Independent analysis of the exome data of family IHG25194 did not yield any nonsense or truncating variants other than the above described DDHD2 mutation.
Pedigrees and mutations. (a) Pedigrees and sequence traces of the DDHD2 families. The c.859C>T mutation segregates in family THI26003 and leads to the formation of a preterminal stop signal at codon 287. The c.1982_1983delAT mutation segregates in family IHG25194; it results in a frameshift at amino acid position 661. (b) Schematic of the DDHD2 gene. The DDHD2 gene contains three known protein domains. The WWE domain is predicted to mediate protein interactions in ubiquitin and ADP ribose conjugation systems. The tandem SAM (sterile alpha motif domain) – DDHD domain is required for phosphoinositide binding.14 Integrity of the family-defining DDHD domain, present in DDHD2 as well as its homolog DDHD1, is necessary for the PLA1 catalytic activity and homo-oligomerization of DDHD2. Catalytic function as well as a positively charged cluster in the SAM domain (Arg434-Lys435-Lys436) also required for phosphoinositide binding are necessary to promote membrane localization.11, 14 Mutations previously described28 are indicated in black, novel mutations in red.
In none of the other four candidate genes additional variants were found in the remaining autosomal recessive HSP families.
Both DDHD2 mutations will destroy the integrity of the DDHD domain and thereby affect catalytic function, membrane localization, phosphoinositide binding and homo-oligomerization (Figure 1). Both mutations therefore likely lead to a complete loss of phospholipase DDHD2 function.
Clinical description
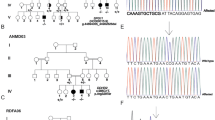
The two siblings of family THI26003 originating from Azerbaijan (Iran) from a consanguineous family background (first degree cousins) developed a progressive spastic gait disorder since early childhood. At the time of examination (disease duration 22–13 years) they were still able to walk unsupported. Due to mental retardation both had attended a special school for mentally disabled children and were working in a sheltered workshop. Spastic paraplegia was further complicated by short stature, high arched palate and dysgenesis of the corpus callosum especially in the dorsal parts (Figures 2a and b).
Cranial MRI/CT scans of DDHD2 patients. MRI/CT scans of DDHD2 patients show dysgenesis of the corpus callosum as well as some paucity of the periventricular white matter (d). (a): coronal MRI (T1 inversion recovery) of THI26003-4. (b): axial CT-scan of THI26003-3. (c): sagittal MRI (T1) of IHG25194-3. (d): axial MRI (T1) of IHG25194-3.
The two brothers of family IHG25194 were of Indian origin and had spastic paraplegia from early childhood. No consanguinity was reported, but both parents originated from the same village and were part of the Muslim community, explaining a possible founder effect. Additional clinical signs and symptoms in both the siblings included mental retardation, mild facial dysmorphism, short stature and dysgenesis of the corpus callosum (Figures 2c and d).
Considering the usually high phenotypic variability in HSP, the phenotype between the two unrelated DDHD2 families is astonishingly similar. Key features of DDHD2-related HSP appear to be spastic paraplegia, mental retardation, short stature and dysgenesis of the corpus callosum (Table 1).
Discussion
Phospholipids are a key component of biological membranes. They are metabolized by the large family of phospholipases that can be classified according to their site of cleavage. The phospholipase A1 family, consisting of extracellular and intracellular enzymes, hydrolyzes the ester bond at the sn-1 position of phospholipids, producing 2-acyl-lysophospholipids and fatty acids. In contrast to most eukaryotic organisms, mammals have three different intracellular phospholipase A1s: phosphatidic acid preferring phospholipase A1 (PA-PLA1/iPLA1α; DDHD1), the SEC23-interacting protein p125 (iPLA1β; SEC23IP) and phospholipase DDHD2 (iPLA1ψ; DDHD2).
The biological function of phospholipase DDHD2 is not fully understood. DDHD2 is ubiquitously expressed; a cytosolic and a membrane-associated pool, localizing to the cis-Golgi and the ER-Golgi intermediate compartment (ERGIC) are in a dynamic equilibrium.11, 12, 13 Overexpression of DDHD2 leads to dispersion of the Golgi and enlargement of the perinuclear ERGIC.12, 14 Conflicting data exists about the effect of DDHD2 depletion. Morikawa et al13 report a specific defect of retrograde transport from the Golgi to the ER; this defect is not confirmed by Sato et al11 who instead postulate a anterograde transport defect from the Golgi to the plasma membrane. The authors contribute these differences to their use of different oligonucleotides for the knockdown of DDHD2 in HeLa cells and possible off-target effects. In spite of these contradictions, the role of DDHD2 in intracellular membrane trafficking is further supported by its homology to another member of the PLA1 family – p125 – that has been shown to be involved in vesicular transport from the ER to the Golgi by interacting with Sec23p, a component of the COP II complex.15
Changes in morphology and dynamics of Golgi and ER functionally connect several subtypes of HSP: Atlastin-1 (SPG3), the CNS expressed member of the atlastin family of GTPases, localizes predominantly to the tubular ER and to a lesser extent to the ERGIC and the cis-Golgi. It is required for the formation of three-way junctions of the ER.16 Receptor expression-enhancing proteins (REEPs) and reticulons form large oligomeric complexes in the ER membrane that are involved in curving the ER membrane, thereby forming the characteristic tubular structure of the smooth ER. Mutations in REEP1 (SPG31) as well as reticulon 2 (RTN2, SPG12) genes cause autosomal dominant forms of HSP.17 Point mutations in BSCL2, causing SPG17, lead to upregulation of ER stress markers.18 The long isoform of spastin mutated in the most common autosomal dominant subtype of HSP (SPAST, SPG4) forms a protein complex with atlastin-1 and REEP1 in the tubular ER network and is thought to coordinate microtubule regulation and membrane modeling.2, 17 The identification of mutations in the gene encoding DDHD2, another protein acting at the ER-to-Golgi interface therefore comes as no surprise.
In addition to its involvement in membrane trafficking, DDHD2 has been shown to possess phospholipase A1 catalytic activity towards phosphatidic acid and other phospholipids,12, 19 a function shared also by DDHD1. Phosphatidic acid has diverse biological functions. It is a precursor for the biosynthesis of triacylglycerols and phosphoglycerols. In addition to its structural functions in biological membranes,20 it is also involved in lipid signaling.21 This phospholipase A1 catalytic activity puts DDHD2 into context with a number of recently discovered HSP genes involved in lipid metabolism. Both seipin (SPG17)22 and spartin (SPG20)23 are involved in formation and regulation of lipid droplets. These dynamic organelles consist of a core of neutral lipids that are covered by a monolayer of amphiphatic lipids containing cholesterol and phospholipids. Phosphatidic acid, preferred substrate for DDHD1 and DDHD2, is essential for lipid droplet assembly.24 The acetyl-CoA transporter SLC33A1 located in the ER membrane and required for the formation of O-acetylated gangliosides is mutated in SPG42. Defects in the metabolism of complex lipids cause at least four more HSP subtypes: in SPG39 the deacetylation of phosphatidylcholine, the major membrane phospholipid, is defective due to mutations in phospholipase B/neuropathy target esterase (PNPLA6),7 mutations in fatty acid-2 hydroxylase (FA2H) affect synthesis of 2-hydroxysphingolipids in SPG3525 and CYP2U1 mutations in SPG49 lead to disturbed ω- and w-1 fatty acid hydroxylation.3 In GBA2, mutated in autosomal recessive HSP SPG46, the conversion of glucosylceramide to free glucose and ceramide by the non-lysosomal glucosylceramidase is deficient.6 In SPG5 mutations in the 7α-hydroxylase gene CYP7B1 not only lead to accumulation of oxysterols in SPG5 patients26 but may also influence neurosteroid metabolism.27
During preparation of this manuscript Schuurs-Hoeijmakers et al28 reported mutations in DDHD2 in four families with autosomal recessive HSP. The phenotype in these families is strikingly similar to the clinical features reported in families THI26003 and IHG25194 with early onset spasticity, mental retardation and TCC being the overlap between all affected family members. The seven DDHD2 mutations described in Schuurs-Hoeijmakers et al28 and our study comprise four frameshift, two nonsense and only one missense mutation so far (Figure 1). Of note, most mutations cluster in the DDHD domain, located in the C-terminal half of the protein. Further studies will have to clarify whether a toxic gain of function of potentially expressed truncated or mutant DDHD2 protein contributes to the phenotype. The nonsense mutation Arg287* that we identified in the Iranian family THI26003 has been described by Schuurs-Hoeijmakers et al28 in another Iranian family; a founder effect is therefore possible.
The identification of mutations in DDHD2, which is involved in Golgi-/ER membrane trafficking and lipid metabolism further demonstrates the critical roles of these essential cellular processes in motor neuron function and helps to understand the molecular mechanisms underlying the pathogenesis of HSPs.
References
Schule R, Schols L : Genetics of hereditary spastic paraplegias. Semin Neurol 2011; 31: 484–493.
Blackstone C : Cellular pathways of hereditary spastic paraplegia. Annu Rev Neurosci 2012; 35: 25–47.
Tesson C, Nawara M, Salih MA et al: Alteration of Fatty-Acid-metabolizing enzymes affects mitochondrial form and function in hereditary spastic paraplegia. Am J Hum Genet 2012; 91: 1051–1064.
Tsaousidou MK, Ouahchi K, Warner TT et al: Sequence alterations within CYP7B1 implicate defective cholesterol homeostasis in motor-neuron degeneration. Am J Hum Genet 2008; 82: 510–515.
Dick KJ, Eckhardt M, Paisan-Ruiz C et al: Mutation of FA2H underlies a complicated form of hereditary spastic paraplegia (SPG35). Hum Mutat 2010; 31: E1251–E1260.
Martin E, Schule R, Smets K et al: Loss of function of glucocerebrosidase GBA2 is responsible for motor neuron defects in hereditary spastic paraplegia. Am J Hum Genet 2013; 92: 238–244.
Rainier S, Bui M, Mark E et al: Neuropathy target esterase gene mutations cause motor neuron disease. Am J Hum Genet 2008; 82: 780–785.
Li H, Durbin R : Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010; 26: 589–595.
McKenna A, Hanna M, Banks E et al: The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010; 20: 1297–1303.
Gonzalez M, Acosta Lebrigio R, van Booven DJ et al: GEnomes Management Application (GEM.app): A new web tool for large-scale collaborative genome analysis. Hum Mutat 2013, (in press).
Sato S, Inoue H, Kogure T, Tagaya M, Tani K : Golgi-localized KIAA0725p regulates membrane trafficking from the Golgi apparatus to the plasma membrane in mammalian cells. FEBS Lett 2010; 584: 4389–4395.
Nakajima K, Sonoda H, Mizoguchi T et al: A novel phospholipase A1 with sequence homology to a mammalian Sec23p-interacting protein, p125. J Biol Chem 2002; 277: 11329–11335.
Morikawa RK, Aoki J, Kano F et al: Intracellular phospholipase A1gamma (iPLA1gamma) is a novel factor involved in coat protein complex I- and Rab6-independent retrograde transport between the endoplasmic reticulum and the golgi complex. J Biol Chem 2009; 284: 26620–26630.
Inoue H, Baba T, Sato S et al: Roles of SAM and DDHD domains in mammalian intracellular phospholipase A(1) KIAA0725p. Biochim Biophys Acta 2012; 1823: 930–939.
Shimoi W, Ezawa I, Nakamoto K et al: p125 is localized in endoplasmic reticulum exit sites and involved in their organization. J Biol Chem 2005; 280: 10141–10148.
Rismanchi N, Soderblom C, Stadler J, Zhu PP, Blackstone C : Atlastin GTPases are required for Golgi apparatus and ER morphogenesis. Hum Mol Genet 2008; 17: 1591–1604.
Park SH, Zhu PP, Parker RL, Blackstone C : Hereditary spastic paraplegia proteins REEP1, spastin, and atlastin-1 coordinate microtubule interactions with the tubular ER network. J Clin Invest 2010; 120: 1097–1110.
Yagi T, Ito D, Nihei Y, Ishihara T, Suzuki N : N88S seipin mutant transgenic mice develop features of seipinopathy/BSCL2-related motor neuron disease via endoplasmic reticulum stress. Hum Mol Genet 2011; 20: 3831–3840.
Tani K, Mizoguchi T, Iwamatsu A, Hatsuzawa K, Tagaya M : p125 is a novel mammalian Sec23p-interacting protein with structural similarity to phospholipid-modifying proteins. J Biol Chem 1999; 274: 20505–20512.
Kooijman EE, Chupin V, de Kruijff B, Burger KN : Modulation of membrane curvature by phosphatidic acid and lysophosphatidic acid. Traffic 2003; 4: 162–174.
Wang X, Devaiah SP, Zhang W, Welti R : Signaling functions of phosphatidic acid. Prog Lipid Res 2006; 45: 250–278.
Boutet E, El Mourabit H, Prot M et al: Seipin deficiency alters fatty acid Delta9 desaturation and lipid droplet formation in Berardinelli-Seip congenital lipodystrophy. Biochimie 2009; 91: 796–803.
Eastman SW, Yassaee M, Bieniasz PD : A role for ubiquitin ligases and Spartin/SPG20 in lipid droplet turnover. J Cell Biol 2009; 184: 881–894.
Fei W, Shui G, Zhang Y et al: A role for phosphatidic acid in the formation of "supersized" lipid droplets. PLoS Genet 2011; 7: e1002201.
Hama H : Fatty acid 2-Hydroxylation in mammalian sphingolipid biology. Biochim Biophys Acta 2010; 1801: 405–414.
Schule R, Siddique T, Deng HX et al: Marked accumulation of 27-hydroxycholesterol in SPG5 patients with hereditary spastic paresis. J Lipid Res 2010; 51: 819–823.
Fex Svenningsen A, Wicher G, Lundqvist J, Pettersson H, Corell M, Norlin M : Effects on DHEA levels by estrogen in rat astrocytes and CNS co-cultures via the regulation of CYP7B1-mediated metabolism. Neurochem Int 2011; 58: 620–624.
Schuurs-Hoeijmakers JH, Geraghty MT, Kamsteeg EJ et al: Mutations in DDHD2, Encoding an Intracellular Phospholipase A(1), Cause a Recessive Form of Complex Hereditary Spastic Paraplegia. Am J Hum Genet 2012; 91: 1073–1081.
Acknowledgements
This study was supported by the National Institute of Health (NIH) (grants 5R01NS072248, 1R01NS075764, 5R01NS054132 to SZ), the Interdisciplinary Center for Clincial Research IZKF Tübingen (grant 1970-0-0 to RS) and the German HSP-Selbsthilfegruppe eV (grant to RS and LS). We thank Dr Tobias Lindig for his critical comments on the neuroradiological findings.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Gonzalez, M., Nampoothiri, S., Kornblum, C. et al. Mutations in phospholipase DDHD2 cause autosomal recessive hereditary spastic paraplegia (SPG54). Eur J Hum Genet 21, 1214–1218 (2013). https://doi.org/10.1038/ejhg.2013.29
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2013.29
Keywords
This article is cited by
-
Lipolysis: cellular mechanisms for lipid mobilization from fat stores
Nature Metabolism (2021)
-
Defining the clinical-genetic and neuroradiological features in SPG54: description of eight additional cases and nine novel DDHD2 variants
Journal of Neurology (2019)
-
Loss of DDHD2, whose mutation causes spastic paraplegia, promotes reactive oxygen species generation and apoptosis
Cell Death & Disease (2018)
-
NT5C2 novel splicing variant expands the phenotypic spectrum of Spastic Paraplegia (SPG45): case report of a new member of thin corpus callosum SPG-Subgroup
BMC Medical Genetics (2017)
-
Spastic paraplegia-linked phospholipase PAPLA1 is necessary for development, reproduction, and energy metabolism in Drosophila
Scientific Reports (2017)