Abstract
Eukaryotic cells utilize the ubiquitin (Ub) system for maintaining a balanced functioning of cellular pathways. Although the Ub system is exclusive to eukaryotes, prokaryotic bacteria have developed an armory of Ub ligase enzymes that are capable of employing the Ub systems of various hosts, ranging from plant to animal cells. These enzymes have been acquired through the evolution and can be classified into three main classes, RING (really interesting new gene), HECT (homologous to the E6-AP carboxyl terminus) and NEL (novel E3 ligases). In this review we describe the roles played by different classes of bacterial Ub ligases in infection and pathogenicity. We also provide an overview of the different mechanisms by which bacteria mimic specific components of the host Ub system and outline the gaps in our current understanding of their functions. Additionally, we discuss approaches and experimental tools for validating this class of enzymes as potential novel antibacterial therapy targets.
Similar content being viewed by others
Introduction
Throughout evolution, prokaryotes have acquired an intricate array of molecular armory that facilitates the hijacking of host cells. In fact, a large portion of the bacterial genome is dedicated for mediating host infection — a central task in bacterial life cycle. The molecular details of this complicated process are now rapidly unfolding and clearly indicate the dependence of bacterial infection strategies on a number of 'toxins' or effectors that have various modes of action. Hijacking of the host cellular environment begins with a rapid rearrangement of actin cytoskeleton within the first 10-30 min of infection1. This abrupt change is brought about by bacterial effectors that activate small GTPases regulating the actin cytoskeleton, such as Rac1 and Cdc422,3. As a result, membrane ruffling is enhanced thus mediating the increased internalization of bacteria via macropinocytosis. Invading bacteria take tight control of the activity of these cellular GTPases, as their prolonged activation would interfere with the bacterial life cycle by disturbing the cellular niche4. Once invasion has been successful, bacteria restore the cytoskeletal architecture by inactivating these cellular GTPases5.
In addition to its role in mediating bacterial internalization, macropinocytosis serves as a 'disguise' that helps bacteria to evade the host defence system. Through residing in modified phagosomal compartments, bacteria remain completely unnoticed by the host defence machinery. The phagosomal compartments have been especially well studied in the case of Salmonella infections, and have become known as Salmonella-containing vacuoles (SCVs). Takeover of cellular systems is facilitated by sophisticated mechanisms that coordinate the delivery of effectors into the host cytoplasm. Many Gram-negative plant and animal pathogens harbor secretion machineries, which are highly analogous to injection needles designed to penetrate membrane structures6. Although the impact of different secretion systems on bacterial virulence may vary, an essential role in pathogenicity is often attributed to the type III secretion system (T3SS)7. This complex injection needle consists of more than 40 proteins, many of which are conserved across plant and animal pathogens8,9. One of the best-described organisms in this regard is again Salmonella, which expresses two T3SSs encoded by Salmonella pathogenicity islands (SPI)-1 and -2. While T3SS-1 is required for active invasion, for example, to deliver effectors that mediate actin cytoskeleton rearrangements, expression of T3SS-2 promotes intracellular survival within SCVs. Following the early stages of infection, the T3SS-1 system is generally downregulated and the low pH environment and nutrient availability within SCVs triggers expression of T3SS-210. In some cases, the exchange of the T3SS needles by Salmonella results in SCV membrane perforation and the release of a subpopulation of bacteria into the cytosol allowing their detection by the host defence system. In this case, the first line of host defence against infection is restriction of bacterial replication via a type of macroautophagy known as xenophagy10. Through this process intracellular bacteria that are not encapsulated in SCVs are sequestered in autophagosomes and their growth is further suppressed by degradation in the hydrolytic environment following lysosomal fusion. In the case of Salmonella, approximately one third of intracellular bacteria are targeted by autophagy one hour post infection11. Salmonella, however, does not need to be free in the cytosol for autophagic degradation to occur. In some instances, perforated vacuoles can also be targeted following binding to cellular galectins, which normally monitor endosomal and lysosomal integrity, resulting in recruitment of the autophagic machinery12. Interestingly, recent studies demonstrate that bacteria have evolved evasive techniques allowing them to escape autophagy and repair SCV membranes damaged by the T3SS-1 needle, therefore restoring the acidic environment within the vacuole and enabling expression of T3SS-2 and subsequent survival13.
Following T3SS-2 expression, bacterial cells are able to deliver the second wave of effectors that hijack signaling cascades of the host to promote invasion and dissemination of bacteria. Intriguingly, among the effectors that are injected by pathogens to the host cellular environment are proteins with ubiquitin (Ub) ligase activity14. Cellular ubiquitylation is proven to be one of the most elaborate post-translational regulation mechanisms developed by eukaryotic cells. It involves a coordinated action of at least three enzymes (E): Ub-activating E1, Ub-conjugating E2 and E3 Ub ligases15. There are two major types of E3 Ub ligases: (1) the RING type that function as scaffolds bringing E2s and the targeted substrates into proximity and (2) the HECT type that form a thioester intermediate with Ub before transferring it to the substrate16. This large class of enzymes plays a key role in ubiquitylation cascades by catalyzing the formation of iso-peptide bonds between the carboxyl terminus of Ub and target lysine (K) residues on the substrate protein15. Ub itself contains seven K residues, all of which could be used to form Ub chains in vivo17. Ub can also form linear chains, in which the carboxyl terminus of one Ub molecule forms a peptide bond with the amino terminus of another Ub18. In fact, distinct structural features of different poly-Ub chains define the recognition of ubiquitylated substrates by specific cellular receptors19. As a consequence, modification of cellular proteins by Ub signals is associated with varied physiological outcomes. For example, substrates modified by K48-linked chains are degraded by the proteasome, K63-linked chains, however, can play a signaling role in recruitment of DNA repair proteins, whereas linear Ub chains regulate the canonical nuclear factor (NF)-κB pathway that activates cytokine gene expression and the host inflammatory response15,17,18,20.
Prokaryotic bacteria do not possess genes encoding Ub. Nevertheless, some bacteria, such as Mycobacterium tuberculosis, harbor proteasomes and degrade their own protein substrates21. It is established that Mycobacterium possesses a functional analogue of the Ub system known as the prokaryotic Ub-like protein (Pup) system22. During pupylation, the Pup protein is covalently conjugated to K residues of protein substrates in a manner similar to Ub23,24. However, unlike the Ub system, which can mediate different physiological outcomes depending on the nature of ubiquitylation, the fate of pupylated substrates is restricted to proteasomal degradation. This is believed to be due to the highly unstructured nature of the Pup protein with a significant part of the protein lacking a stable secondary or tertiary structure25,26,27. Therefore, despite the overall functional similarity between the Pup and Ub systems, the intrinsically structurally disordered Pup may explain the inability of bacterial pathogens to utilize their Pup system as an effective modifier for manipulation of host signaling pathways. It is therefore likely that bacterial cells have evolved alternative routes, including the expression and release of Ub ligase-like effectors that are more compatible with the hosts' Ub machinery. As a consequence, bacteria are able to hijack the hosts' own Ub system, thereby manipulating a multitude of signaling cascades to promote bacterial survival.
In recent years it has become evident that bacterial Ub ligase-like effectors are central to the bacterial life cycle. Therefore, multiple studies have focused on identifying effector-specific host substrates in order to decipher key host signaling cascade modulations that are essential for bacterial survival and dissemination. To provide a better understanding of host-pathogen interactions, in this review we focus on known bacterial Ub ligases from different pathogens and describe their roles in modulating signaling cascades of the host. We also discuss the possibility of targeting these effectors for combating bacterial infections.
Ub ligase mimicry in the 'arms race' between pathogen and host
Pseudomonas syringae is a pathogen of tomato and Arabidopsis thaliana cells and dedicates about 7% of its genome for producing effectors that dampen host innate immunity and promote disease in plants28. Plant cells, in turn, recognize the bacterial effectors released to the cellular environment through a process known as effector-triggered immunity (ETI)29. This type of response to infection leads to a localized immunity-associated programmed cell death (PCD), in which plants 'sacrifice' a limited portion of the leaf to protect the rest of the plant from a more severe systemic infection30. The interaction between Pseudomonas and its host involves a co-evolution of virulence effectors and the ETI pathway, which in many aspects could be depicted as a type of 'arms race'. Not surprisingly, bacterial Ub ligase-like effectors are at the heart of this battle. For example, the Pseudomonas AvrPtoB effector represents a mechanism by which bacteria suppress this conserved ETI pathway through inhibiting PCD activators29. The carboxy-terminal domain (CTD) of AvrPtoB is essential to avoid recognition by plant immunity, as deletion of this domain allows the amino-terminal region of AvrPtoB to be detected by certain tomato varieties leading to immunity-associated PCD31. The function and activity of the AvrPtoB CTD could not be predicted from initial examination of the primary amino acid sequence. It was only when Janjusevic and colleagues revealed the structure of the AvrPtoB CTD that it became apparent that it has a remarkable resemblance to the canonical eukaryote-specific RING domain present in Ub ligases32. This highly homologous domain serves as a binding site for E2 enzymes as mutations affecting this domain typically block E2 binding and impair Ub ligase activity33. Therefore, the presence of this conserved RING domain allows AvrPtoB to hijack the host Ub system for its own purposes, which is essential for bacterial virulence31,32.
Through recent molecular studies it has become apparent how host resistance components of the plant immune system enhance the capacity to recognize and respond to Pseudomonas infection. The Fen kinase directly interacts with the AvrPtoB amino terminus and is responsible for activating the plant ETI. Interestingly, the RING domain located in the CTD of AvrPtoB facilitates ubiquitylation and degradation of the Fen kinase, thereby counteracting ETI and increasing susceptibility of plants to disease34,35. Remarkably, the AvrPtoB-induced degradation of the Fen kinase is mediated through the host's own proteasomal system. Thus, in the case of the Fen kinase, by acquiring a Ub ligase activity, AvrPtoB has thwarted a highly conserved host resistance mechanism via 'turning' the host's proteasomal system against the host.
The 'arms race' between the host and the pathogen is further demonstrated by the interaction between AvrPtoB and another host target — the Pto kinase. Unlike the Fen kinase, this interaction favors the host defence system. By phosphorylating a regulatory threonine residue within the catalytic cleft of the RING domain of AvrPtoB, Pto diminishes the ligase activity of AvrPtoB and reduces ubiquitylation of its targets, including Fen — conferring Pto-mediated resistance against Pseudomonas infection35. Therefore, the outcome of Pseudomonas infection depends on a competition between the phosphorylation and ubiquitylation activities mediated by the host kinase and the invading effector ligase, respectively. Pto represents a host variant molecule with higher kinase activity and hence this defence countermeasure grants the host the ability to fight back against the pathogen.
Ub ligase mimicry extends to mammalian pathogens
Plant pathogens are not unique in their ability to develop effectors that structurally or functionally mimic host Ub ligases to achieve their goals. Many human pathogens also utilize Ub ligase-like effectors that are secreted to the host cells (Table 1). One such pathogen, Salmonella enterica, can cause salmonellosis in humans, a disease commonly acquired by ingestion of contaminated food or water. Clinical syndromes of this infection include severe typhoid fever, caused by Salmonella typhi and paratyphi species, with fatality rate ranging between 10% and 20% in untreated cases and 1% following treatment with appropriate antibiotics. Additionally, a wide range of milder clinical syndromes have been described for non-typhoid Salmonella, which in healthy adults causes diarrhoeal disease with an untreated case fatality rate of 0.1%36.
Salmonella developed sophisticated mechanisms for effective invasion of macrophages and epithelial cells. Among the effectors that Salmonella delivers to the host are enzymes that possess Ub ligase activity and are implicated in suppressing the inflammatory host response14. One such effector is the SopA E3-like enzyme. Despite the lack of sequence similarity, the overall molecular structure of the SopA carboxyl terminus is highly reminiscent of the eukaryotic HECT domain with its characteristic architecture containing a two-lobe structure37. Much like host HECT Ub ligases, SopA utilizes a catalytic cysteine residue to form a Ub thioester intermediate, which is essential for its in vitro ubiquitylation activity38,39. Although there are no reported substrates to date, SopA is known to play a key role in the induction of enteritis. Intriguingly, both sopA insertion and deletion mutants demonstrate a 5-fold decrease in polymorphonuclear neutrophil transepithelial migration, a hallmark of Salmonella infection, highlighting the importance of SopA in mediating Salmonella pathogenicity40.
SopA shares 26% similarity with the NleL effector from enterohaemorrhagic and enteropathogenic E. coli strains (EHEC and EPEC, respectively)38,41. Similar to SopA, NleL is also delivered to the host's cytoplasm through the T3SS and has Ub ligase activity38,42,43. Importantly, the formation of attaching and effacing lesions, a hallmark of pathogenic E. coli infection following bacterial attachment to epithelial cells, is dependent on NleL42. Both SopA and NleL bacterial Ub ligases interact with host E2s in a manner similar to host E3s. By utilizing hydrophobic interactions between the conserved phenylalanine in the host UbcH7 E2 enzyme and the aromatic residues in SopA and NleL, these bacterial Ub ligase-like enzymes occupy the same region on the E2 as the host HECT E3s. This indicates that bacterial effectors may also interfere with cellular ubiquitylation by limiting the availability of cellular E2 enzymes.
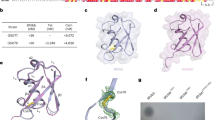
Previous studies of human HECT ligases revealed that amino-terminal extensions upstream of the HECT domain determine substrate recognition. For this purpose, human HECT Ub ligases contain a variety of domains that facilitate substrate recognition and protein-protein interactions44. The domain architecture of bacterial SopA and NleL enzymes is also organized in a similar fashion. The HECT-like domain is located at the carboxyl terminus with the amino-terminal part of SopA and NleL containing a parallel β-helix domain that is hypothesized to serve as a substrate-binding region38,39 (Figure 1). Interestingly, the C-lobes of both SopA and NleL HECT-like domains possess a high degree of conformational flexibility, indicating that bacterial Ub ligases harbor the necessary structural organization that allows them to facilitate Ub transfer from E2s onto the targeted substrates, similarly to eukaryotic HECT E3s39.
Comparison of the NleL and SopA crystal structures. Ribbon representations of the NleL (PDB code: 3NB2) and SopA (PDB code: 2QYU) HECT-like bacterial Ub ligases. Catalytic cysteine residues of NleL (C753) and SopA (C753) are represented as red spheres. The amino (N) and carboxyl (C) lobes of the HECT domain are indicated, as well as the amino-terminal substrate recognition β-helix domain.
As indicated, SopA and NleL are crucial for mediating Salmonella-induced enteritis and E. coli lesions, respectively. This calls for the identification of their specific host targets, which is promising to unveil key host pathways modulated by these Ub ligases involved in enhancing bacterial pathogenicity. This information, in addition to increasing our understanding of host-pathogen interactions, can also provide insight into novel therapeutic approaches, as discussed later, to combat bacterial infection.
Among the largest groups of bacterial effectors is the family of NleG proteins that are present in a variety of pathogenic EHEC and EPEC strains. NleG effectors are secreted via the T3SS system and have been linked to lethal E. coli infections43,45. Comparative sequence analysis of NleG family members revealed a highly conserved region of ∼100 amino acids in the CTD, which is annotated as the domain of unknown function (DUF) 1076. Interestingly, the NMR structure of NleG2-3 CTD, an NleG family member, uncovered the presence of a motif similar to the eukaryotic RING/U-box domain, which is a modified version of the RING domain that confers Ub ligase activity46,47,48. Almost all NleG effectors contain this conserved motif embedded in the sequence of their CTDs, suggesting that they function through host Ub ligase mimicry to enhance pathogenicity (Figure 2A)49,50. In fact, in vitro Ub ligase activity has been reported for some family members, including NleG2-3, NleG5-1, NleG6-2 and NleG948. Remarkably, the tertiary structure of the NleG2-3 CTD is strikingly similar to that of the P. syringae AvrPtoB CTD, despite low amino acid sequence identity51 (Figure 2B). This provides additional evidence supporting a potential role of NleGs as effectors with Ub ligase activity inside the host cellular environment.
The carboxyl terminus of NleG effectors harbors RING/U-box domain. (A) Amino acid sequence alignment of NleG family member carboxy-terminal domains as analyzed by Clustal Omega49,50. Conserved amino acids corresponding to the RING/U-box motif are denoted with asterisks. NleG family members with demonstrated Ub ligase activity in vitro are highlighted in bold italic48. (B) Comparison of RING/U-box-like domains of AvrPtoB and NleG2-3. CTD structures of AvrPtoB (PDB code: 2FD4) and NleG2-3 (PDB code: 2KKY) were superimposed using Dalilite server51. For clarity in comparison, peripheral elements of the CTDs are removed from both structures, showing only the core of the RING/U-box domains.
In contrast to the highly conserved CTDs, the amino termini of NleGs have low sequence identity and do not harbor any functional domains that can hint on how the cellular targets are recognized. Attempts to identify protein-protein interactions of some NleG effectors using the yeast two-hybrid (Y2H) technology were unsuccessful, despite finding novel high-confidence interacting partners of other effectors of pathogenic E. coli in the same screen52. Therefore, at present, mechanisms by which NleGs modulate host signaling are not well understood.
Legionella pneumophila utilizes yet another strategy that is based on mimicking functional domains involved in host Ub signaling. Legionella pneumophila invades and establishes a replicative niche in human macrophages by residing in Legionella-containing vacuoles (LCVs)53. In addition to delivering effectors containing U-box motifs that hijack host E2s to mediate target ubiquitylation54,55, Legionella also injects a family of F-box-containing proteins56. By utilizing the conserved F-box motif, these effectors interact with the Skp1 subunit of host multimeric Ub ligase complexes known as SCF (Skp1-Cullin-F-box) and act as adaptors that can recruit substrates for ubiquitylation56. For example, the AnkB effector of Legionella is anchored to the cytosolic face of the LCVs, where this effector mediates K48-specific ubiquitylation of proteins through the host's SCF complex56,57,58. By inducing degradation of proteins en mass AnkB satisfies the high demands of Legionella for amino acids and thus acts as a crucial weapon for promoting intracellular growth of this pathogen57.
Bacteria further diversify the armory of Ub ligases
Structural or functional mimicry of the host Ub system has apparently proven to be an effective strategy for manipulating host signaling. In addition to acquiring effectors with RING and HECT-like domains, bacterial pathogens further diversified the armory of Ub ligases by evolving enzymes that are structurally distinct from their mammalian counterparts, termed NELs (novel E3 ligases). NELs constitute a family of at least 15 enzymes that spans across 6 pathogenic bacterial genera, including highly pathogenic Salmonella and Shigella59. NELs harbor three essential E3 ligase elements. First, the E2-interacting surface residues are involved in hijacking host E2s charged with Ub. Second, although NELs lack the bona fide bi-lobal HECT domain, these bacterial ligases have a catalytic cysteine residue utilized for Ub transfer. Last, the presence of leucine-rich repeats (LRRs) of variable length ensures the recognition of a wide array of targets (Figure 3)60,61,62,63,64.
Structures of NELs from various pathogens. Ribbon representations of full-length NEL proteins (SspH2, SlrP, YopM, IpaH3), LRR (SspH1) and NEL (IpaH1.4) domains from various pathogens. Salmonella enteric SlrP and SspH1 structures are depicted in substrate-bound states. Catalytic cysteine residues of SspH1 (C580), SlrP (C546), IpaH3 (C363) and IpaH1.4 (C368) are represented as red spheres.
Shigella flexneri IpaH effectors were the first described NELs. Shigella encodes at least three IpaH ligase effectors on its virulence plasmids and seven additional effectors are encoded by its genomic DNA65,66,67,68. All of IpaH effector proteins are secreted via the T3SS and thus potentially play a role in pathogenicity69. The IpaH9.8 ligase (encoded by a 9.8 kb DNA fragment on the virulence plasmid) was found to disrupt the pheromone response through supressing the MAPK pathway by promoting proteasome-dependent degradation of the MAPK kinase Ste7 when overexpressed in budding yeast59. Although IpaH9.8 E3 Ub ligase activity has been reported in vitro, it is still not clear whether IpaH9.8 affects the MAPK pathway in mammalian cells in a similar manner. IpaH9.8 also attenuates NF-κB signaling by degrading NEMO (NF-κB essential modulator), an essential component of this signaling cascade70. Mice infected with Shigella expressing a catalytically inactive IpaH9.8 C337A mutant exhibited reduced bacterial multiplication with an augmented inflammatory response as compared with mice infected with the wild-type strain70. Another member of the IpaH ligase family, IpaH7.8, was also reported to have Ub ligase activity in vitro and has been shown to facilitate the escape of virulent bacteria from endocytic vacuoles in mouse and human macrophages59,71. Control mice infected with wild-type Shigella exhibited enhanced bacterial multiplication as compared to mice infected with mutant bacteria lacking IpaH7.8. IpaH7.8 was also suggested to play a central role in promoting macrophage cell death through the activation of inflammasomes. Glomulin, a natural inflammasome inhibitor, is targeted by IpaH7.8 for degradation, which in turn stimulates inflammasome activation, macrophage cell death and dissemination of bacteria72. Another example of the role of the IpaH family in modulating the host's inflammatory pathways is IpaH0722 that was demonstrated to ubiquitylate and mediate the proteasomal degradation of TRAF2, thereby inhibiting protein kinase C-mediated NF-κB pathway activation upon Shigella infection73. Additionally, in a Y2H screen the NEL IpaH4.5 was identified to interact with the NF-κB transcription factor subunit p65 and was shown to drive p65 ubiquitylation and to inhibit TNFα-mediated NF-κB activation in vitro74.
In addition to the NELs described above, the role of NELs in promoting bacterial pathogenicity is further demonstrated in Salmonella infection. Salmonella encodes three members of the NEL family: SspH1, SspH2 and SlrP. All three members were shown to contribute to virulence in animal models of infection by mechanisms that are currently not well understood. SlrP mutant bacteria show colonization defects upon infection of mice, whereas both SspH1 and SspH2 are required for the induction of lethal infections in calves75,76. Using a Y2H screening approach, both Thioredoxin-1 and the host chaperone ERdJ3 were identified as interacting proteins of SlrP77,78. These data implicate SlrP in the inhibition of the overall stress response upon bacterial infection, yet more studies are required to dissect the exact mode of action of SlrP.
Once inside the host cell, SspH1 localizes to the nucleus, inhibits NF-κB-dependent gene expression and contributes to regulation of pro-inflammatory cytokine production. The SspH1 LRR domain was shown to bind protein kinase PKN1 in Y2H experiments79 and ubiquitylate PKN1 in vitro59. Initial affinity purification mass spectrometry (AP-MS) efforts identified the NOD receptor co-chaperone Sgt1 as an SspH2-binding partner. Subsequently, SspH2 was also demonstrated to enhance NOD1-mediated responses in cells. Mechanistic studies show that SspH2 stimulates the non-proteolytic ubiquitylation of NOD1 in heterologous expression experiments leading to agonist-independent activation of signaling80,81.
Structural studies provide interesting insight into the regulation of NEL ligase activity64. It appears that in the absence of physiological substrates, the catalytic cysteine residue is structurally occluded by carboxy-terminal LRRs, resulting in autoinhibition of NELs. This autoinhibitory mechanism is reminiscent of mammalian HECT Ub ligases. For example, autoinhibition of Smurf2 catalytic activity by its C2 domain is suggested to reduce spurious ligase activity82. Although the purpose of NEL autoinhibition is still unclear, it is possible that this regulation is important for restricting their activity to avoid autoubiquitylation and subsequent degradation or simply to minimize detection by the host. NEL autoinhibition might also function as an additional regulatory step to prevent competition with other bacterial ligases for host E2s in situations where NEL host substrates are absent. As with other bacterial Ub ligase-like effectors, the full spectrum of ubiquitylated substrates by NELs is currently unknown. Moreover, a global overview of the ubiquitylated host proteome by these effectors and knowledge on the dynamics of substrate ubiquitylation during infection are currently lacking.
In addition to Shigella and Salmonella species, pathogenic Yersinia species also encode the YopM effector that is required for full virulence in mouse infection models83. Although YopM contains a number of LRRs in its carboxyl terminus and its amino terminus was suggested to harbor similarity to the NEL domain of Salmonella and Shigella effectors, a Ub ligase activity for this effector has not been reported84,85. Instead, YopM serves as a scaffolding protein that facilitates the formation of a novel complex between RSK and PRK host kinases that normally do not interact86. Although this results in stimulation of kinase activity, at present the molecular role of the YopM/RSK/PRK complex remains unknown87. YopM also directly inhibits caspase-1 function in infected macrophages, which is essential for Yersinia pseudotuberculosis virulence in vivo88.
Bacterial Ub ligase-like effectors as potential drug targets
The examples of bacterial Ub ligase-like effectors described above illustrate an important role of these enzymes in modulating host cellular signaling, thus facilitating infection and dissemination of pathogens. Although much remains to be learnt about different classes of bacterial Ub ligase-like enzymes and their mechanisms of action, it is important to consider the pharmacological inhibition of their activity as a novel strategy for treating severe pathogenic bacterial infections. Ub ligase drug discovery is minimal compared to other classes of signaling molecules, such as protein kinases or G protein-coupled receptors89. This is mainly due to limited tools currently available for developing robust high-throughput screening assays against Ub ligases of interest. The absence of ATP-binding pockets within the RING and HECT catalytic domains presents further challenges associated with targeting protein-protein interactions between Ub ligases and their cognate substrates or upstream E2-conjugating enzymes. Despite these limitations, a number of approaches, as described below, could potentially be utilized for targeting bacterial Ub ligase-like enzymes.
During the ubiquitylation reaction Ub ligases are positioned in close proximity to substrate proteins, which enables the application of the fluorescence resonance energy transfer (FRET) technology90,91. Provided that efforts in identifying high-confidence substrates of bacterial Ub ligase-like effectors will be intensified in the future, one could envision developing robust assays for targeting the interaction between Ub ligases and their cognate substrates utilizing FRET or similar in vitro reconstitution assays92. Structure-driven approaches also offer powerful tools in identifying potent inhibitors of Ub ligases93,94. Therefore, it is important to continue obtaining high-resolution structures of protein-protein interactions between bacterial Ub ligase-like enzymes and their substrates or partner E2s. Substrate-independent in vitro assays have also been designed for developing inhibitors of autocatalytic activity of physiologically relevant Ub ligases95. Since the catalytic activity of NELs plays an important role in the life cycle of a number of human pathogens and the conserved bacterial NEL domain appears distinct from known mammalian Ub ligases, the development of a broad-spectrum 'pan-NEL' inhibitor could potentially offer unmatched therapeutic benefits in multiple disease areas. More recently, pharmacological targeting of Ub ligase activity has been advanced to cellular assays96. Adopting a similar approach might therefore enable the generation of small-molecule inhibitors against the NEL active site within the host cellular environment. Since NELs are unique to bacteria, one could anticipate a very low cross-reactivity of such inhibitors against the host Ub ligases thereby potentially reducing non-specific side effects.
Concluding remarks
The power of Y2H technology in allowing the fast interrogation of binary interactions between bacterial Ub ligase-like effectors and host substrates on a proteome-wide scale was instrumental for the identification of numerous host-pathogen interactions97,98. However, this technology also harbors several limitations, such as the use of yeast as a surrogate organism for mimicking the environment of the mammalian cell and the difficulty in detecting interactions involving multiple binding partners. In this context, AP-MS approaches provide a valuable complementary technology for the analysis of protein interaction networks and may also prove to be essential for the identification of physiological targets of bacterial Ub ligase-like enzymes. Of utmost importance is the development of methodology enabling the identification of proteins that are specifically ubiquitylated upon infection of host cells by various pathogens, which should significantly improve the identification of high-confidence targets that are relevant for bacterial infection. In addition, given that target phosphorylation plays an important role in substrate recognition by Ub ligases99, it is also important to identify host cellular kinases that may participate in target recognition by bacterial E3s. Furthermore, the continuous advancements in our understanding of target selectivity and activity of bacterial Ub ligases-like effectors necessitate the development of novel research tools. Activity-based probes (ABPs) harboring chemical warheads that covalently bind to the active site of enzymes have been previously developed for monitoring the activity of the Ub system100,101. Recent improvements of ABPs made it possible to detect the activity of host Ub ligases102. Therefore, ABPs could potentially represent useful tools for monitoring the activity of bacterial Ub ligase-like enzymes and could also be utilized in pharmacological screening to search for a more effective antibacterial therapy.
In conclusion, given the complexity of the host-pathogen 'arms race', it is instrumental that we combine the power of multiple techniques to tackle the different aspects of host-pathogen interactions. This promises to vastly expand our current knowledge and provide a robust foundation for identifying new tools to combat bacterial infection.
References
Kubori T, Galán JE . Temporal regulation of salmonella virulence effector function by proteasome-dependent protein degradation. Cell 2003; 115:333–342.
Hardt WD, Chen LM, Schuebel KE, Bustelo XR, Galán JE . S. typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell 1998; 93:815–826.
Stender S, Friebel A, Linder S, Rohde M, Mirold S, Hardt WD . Identification of SopE2 from Salmonella typhimurium, a conserved guanine nucleotide exchange factor for Cdc42 of the host cell. Mol Microbiol 2000; 36:1206–1221.
Stebbins CE, Galán JE . Structural mimicry in bacterial virulence. Nature 2001; 412:701–705.
Fu Y, Galán JE . A salmonella protein antagonizes Rac-1 and Cdc42 to mediate host-cell recovery after bacterial invasion. Nature 1999; 401:293–297.
Mueller CA, Broz P, Cornelis GR . The type III secretion system tip complex and translocon. Mol Microbiol 2008; 68:1085–1095.
Galán JE, Wolf-Watz H . Protein delivery into eukaryotic cells by type III secretion machines. Nature 2006; 444:567–573.
Portaliou AG, Tsolis KC, Loos MS, Zorzini V, Economou A . Type III secretion: Building and operating a remarkable nanomachine. Trends Biochem Sci 2016; 41:175–189.
Diepold A, Armitage JP . Type III secretion systems: the bacterial flagellum and the injectisome. Philos Trans R Soc Lond B Biol Sci 2015; 370:20150020.
Cemma M, Brumell JH . Interactions of pathogenic bacteria minireview with autophagy systems. Curr Biol 2012; 22:R540–R545.
Birmingham CL, Smith AC, Bakowski MA, Yoshimori T, Brumell JH . Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J Biol Chem 2006; 281:11374–11383.
Thurston TLM, Wandel MP, Muhlinen von N, Foeglein Á, Randow F . Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature 2012; 482:414–418.
Kreibich S, Emmenlauer M, Fredlund J, et al. Autophagy proteins promote repair of endosomal membranes damaged by the Salmonella type three secretion system 1. Cell Host Microbe 2015; 18:527–537.
Ashida H, Kim M, Sasakawa C . Exploitation of the host ubiquitin system by human bacterial pathogens. Nat Rev Microbiol 2014; 12:399–413.
Capili AD, Lima CD . Taking it step by step: mechanistic insights from structural studies of ubiquitin/ubiquitin-like protein modification pathways. Curr Opin Struct Biol 2007; 17:726–735.
Vittal V, Stewart MD, Brzovic PS, Klevit RE . Regulating the regulators: Recent revelations in the control of E3 ubiquitin ligases. J Biol Chem 2015; 290:21244–21251.
Ikeda F, Dikic I . Atypical ubiquitin chains: new molecular signals. EMBO Rep 2008; 9:536–542.
Walczak H, Iwai K, Dikic I . Generation and physiological roles of linear ubiquitin chains. BMC Biol 2012; 10:23.
Dikic I, Wakatsuki S, Walters KJ . Ubiquitin-binding domains — from structures to functions. Nat Rev Mol Cell Biol 2009; 10:659–671.
Ulrich HD, Walden H . Ubiquitin signalling in DNA replication and repair. Nat Rev Mol Cell Biol 2010; 11:479–489.
Baumeister W, Walz J, Zühl F, Seemüller E . The proteasome: paradigm of a self-compartmentalizing protease. Cell 1998; 92:367–380.
Pearce MJ, Mintseris J, Ferreyra J, Gygi SP, Darwin KH . Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science 2008; 322:1104–1107.
Striebel F, Imkamp F, Sutter M, Steiner M, Mamedov A, Weber-Ban E . Bacterial ubiquitin-like modifier Pup is deamidated and conjugated to substrates by distinct but homologous enzymes. Nat Struct Mol Biol 2009; 16:647–651.
Cerda-Maira FA, McAllister F, Bode NJ, Burns KE, Gygi SP, Darwin KH . Reconstitution of the Mycobacterium tuberculosis pupylation pathway in Escherichia coli. EMBO Rep 2011; 12:863–870.
Wang T, Darwin KH, Li H . Binding-induced folding of prokaryotic ubiquitin-like protein on the Mycobacterium proteasomal ATPase targets substrates for degradation. Nat Struct Mol Biol 2010; 17:1352–1357.
Burns KE, Pearce MJ, Darwin KH . Prokaryotic ubiquitin-like protein provides a two-part degron to Mycobacterium proteasome substrates. J Bacteriol 2010; 192:2933–2935.
Cobbert JD, DeMott C, Majumder S, et al. Caught in action: selecting peptide aptamers against intrinsically disordered proteins in live cells. Sci Rep 2015; 5:9402.
Buell CR, Joardar V, Lindeberg M, et al. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci USA 2003; 100:10181–10186.
Dodds PN, Rathjen JP . Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet 2010; 11:539–548.
Oh CS, Martin GB . Effector-triggered immunity mediated by the Pto kinase. Trends Plant Sci 2011; 16:132–140.
Abramovitch RB, Janjusevic R, Stebbins CE, Martin GB . Type III effector AvrPtoB requires intrinsic E3 ubiquitin ligase activity to suppress plant cell death and immunity. Proc Natl Acad Sci USA 2006; 103:2851–2856.
Janjusevic R, Abramovitch RB, Martin GB, Stebbins CE . A bacterial inhibitor of host programmed cell death defenses is an E3 ubiquitin ligase. Science 2006; 311:222–226.
Zheng N, Schulman BA, Song L, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 2002; 416:703–709.
Rosebrock TR, Zeng L, Brady JJ, Abramovitch RB, Xiao F, Martin GB . A bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity. Nature 2007; 448:370–374.
Ntoukakis V, Mucyn TS, Gimenez-Ibanez S, et al. Host inhibition of a bacterial virulence effector triggers immunity to infection. Science 2009; 324:784–787.
LaRock DL, Chaudhary A, Miller SI . Salmonellae interactions with host processes. Nat Rev Microbiol 2015; 13:191–205.
Lin DY, Diao J, Chen J . Crystal structures of two bacterial HECT-like E3 ligases in complex with a human E2 reveal atomic details of pathogen-host interactions. Proc Natl Acad Sci USA 2012; 109:1925–1930.
Lin DY, Diao J, Zhou D, Chen J . Biochemical and structural studies of a HECT-like ubiquitin ligase from Escherichia coli O157:H7. J Biol Chem 2010; 286:441–449.
Diao J, Zhang Y, Huibregtse JM, Zhou D, Chen J . Crystal structure of SopA, a Salmonella effector protein mimicking a eukaryotic ubiquitin ligase. Nat Struct Mol Biol 2007; 15:65–70.
Wood MW, Jones MA, Watson PR, et al. The secreted effector protein of Salmonella dublin, SopA, is translocated into eukaryotic cells and influences the induction of enteritis. Cell Microbiol 2000; 2:293–303.
Li M, Rosenshine I, Yu HB, et al. Identification and characterization of NleI, a new non-LEE-encoded effector of enteropathogenic Escherichia coli (EPEC). Microbes Infect 2006; 8:2890–2898.
Piscatelli H, Kotkar SA, McBee ME, et al. The EHEC type III effector NleL is an E3 ubiquitin ligase that modulates pedestal formation. PLoS One 2011; 6:e19331.
Tobe T, Beatson SA, Taniguchi H, et al. An extensive repertoire of type III secretion effectors in Escherichia coli O157 and the role of lambdoid phages in their dissemination. Proc Natl Acad Sci USA 2006; 103:14941–14946.
Scheffner M, Kumar S . Mammalian HECT ubiquitin-protein ligases: biological and pathophysiological aspects. Biochim Biophys Acta 2014; 1843:61–74.
Donnenberg MS, Hazen TH, Farag TH, et al. Bacterial factors associated with lethal outcome of enteropathogenic Escherichia coli infection: genomic case-control studies. PLoS Negl Trop Dis 2015; 9:e0003791.
Aravind L, Koonin EV . The U box is a modified RING finger — a common domain in ubiquitination. Curr Biol 2000; 10:R132–R134.
Hatakeyama S, Nakayama K-II . U-box proteins as a new family of ubiquitin ligases. Biochem Biophys Res Commun 2003; 302:635–645.
Wu B, Skarina T, Yee A, et al. NleG type 3 effectors from enterohaemorrhagic Escherichia coli are U-box E3 ubiquitin ligases. PLoS Pathog 2010; 6:e1000960.
Sievers F, Wilm A, Dineen D, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 2011; 7:539.
Goujon M, McWilliam H, Li W, et al. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res 2010; 38:W695–W699.
Hasegawa H, Holm L . Advances and pitfalls of protein structural alignment. Curr Opin Struct Biol 2009; 19:341–348.
Blasche S, Arens S, Ceol A, et al. The EHEC-host interactome reveals novel targets for the translocated intimin receptor. Sci Rep 2014; 4:7531.
Isberg RR, O'Connor TJ, Heidtman M . The Legionella pneumophila replication vacuole: making a cozy niche inside host cells. Nat Rev Microbiol 2009; 7:13–24.
Kubori T, Hyakutake A, Nagai H . Legionella translocates an E3 ubiquitin ligase that has multiple U-boxes with distinct functions. Mol Microbiol 2008; 67:1307–1319.
Quaile AT, Urbanus ML, Stogios PJ, et al. Molecular characterization of LubX: functional divergence of the U-box fold by Legionella pneumophila. Structure 2015; 23:1459–1469.
Ensminger AW, Isberg RR . E3 ubiquitin ligase activity and targeting of BAT3 by multiple Legionella pneumophila translocated substrates. Infect Immun 2010; 78:3905–3919.
Price CTD, Al-Quadan T, Santic M, Rosenshine I, Kwaik YA . Host proteasomal degradation generates amino acids essential for intracellular bacterial growth. Science 2011; 334:1553–1557.
Lomma M, Dervins-Ravault D, Rolando M, et al. The Legionella pneumophila F-box protein Lpp2082 (AnkB) modulates ubiquitination of the host protein parvin B and promotes intracellular replication. Cell Microbiol 2010; 12:1272–1291.
Rohde JR, Breitkreutz A, Chenal A, Sansonetti PJ, Parsot C . Type III secretion effectors of the IpaH family are E3 ubiquitin ligases. Cell Host Microbe 2007; 1:77–83.
Singer AU, Rohde JR, Lam R, et al. Structure of the Shigella T3SS effector IpaH defines a new class of E3 ubiquitin ligases. Nat Struct Mol Biol 2008; 15:1293–1301.
Zhu Y, Li H, Hu L, et al. Structure of a Shigella effector reveals a new class of ubiquitin ligases. Nat Struct Mol Biol 2008; 15:1302–1308.
Zouhir S, Bernal-Bayard J, Cordero Alba M, et al. The structure of the Slrp-Trx1 complex sheds light on the autoinhibition mechanism of the type III secretion system effectors of the NEL family. Biochem J 2014; 464:135–144.
Keszei AFA, Tang X, McCormick C, et al. Structure of an SspH1-PKN1 complex reveals the basis for host substrate recognition and mechanism of activation for a bacterial E3 ubiquitin ligase. Mol Cell Biol 2014; 34:362–373.
Quezada CM, Hicks SW, Galán JE, Stebbins CE . A family of Salmonella virulence factors functions as a distinct class of autoregulated E3 ubiquitin ligases. Proc Natl Acad Sci USA 2009; 106:4864–4869.
Venkatesan MM, Goldberg MB, Rose DJ, Grotbeck EJ, Burland V, Blattner FR . Complete DNA sequence and analysis of the large virulence plasmid of Shigella flexneri. Infect Immun 2001; 69:3271–3285.
Buchrieser C, Glaser P, Rusniok C, et al. The virulence plasmid pWR100 and the repertoire of proteins secreted by the type III secretion apparatus of Shigella flexneri. Mol Microbiol 2000; 38:760–771.
Wei J, Goldberg MB, Burland V, et al. Complete genome sequence and comparative genomics of Shigella flexneri serotype 2a strain 2457T. Infect Immun 2003; 71:2775–2786.
Jin Q, Yuan Z, Xu J, et al. Genome sequence of Shigella flexneri 2a: insights into pathogenicity through comparison with genomes of Escherichia coli K12 and O157. Nucleic Acids Res 2002; 30:4432–4441.
Ashida H, Toyotome T, Nagai T, Sasakawa C . Shigella chromosomal IpaH proteins are secreted via the type III secretion system and act as effectors. Mol Microbiol 2007; 63:680–693.
Ashida H, Kim M, Schmidt-Supprian M, Ma A, Ogawa M, Sasakawa C . A bacterial E3 ubiquitin ligase IpaH9.8 targets NEMO/IKKγ to dampen the host NF-κB-mediated inflammatory response. Nat Cell Biol 2009; 12:66–73.
Fernandez-Prada CM, Hoover DL, Tall BD, Hartman AB, Kopelowitz J, Venkatesan MM . Shigella flexneri IpaH(7.8) facilitates escape of virulent bacteria from the endocytic vacuoles of mouse and human macrophages. Infect Immun 2000; 68:3608–3619.
Suzuki S, Mimuro H, Kim M, et al. Shigella IpaH7.8 E3 ubiquitin ligase targets glomulin and activates inflammasomes to demolish macrophages. Proc Natl Acad Sci USA 2014; 111:E4254–E4263.
Ashida H, Nakano H, Sasakawa C . Shigella IpaH0722 E3 ubiquitin ligase effector targets TRAF2 to inhibit PKC-NF-κB activity in invaded epithelial cells. PLoS Pathog 2013; 9:e1003409.
Wang F, Jiang Z, Li Y, et al. Shigella flexneri T3SS effector IpaH4.5 modulates the host inflammatory response via interaction with NF-κB p65 protein. Cell Microbiol 2012; 15:474–485.
Tsolis RM, Townsend SM, Miao EA, et al. Identification of a putative Salmonella enterica serotype typhimurium host range factor with homology to IpaH and YopM by signature-tagged mutagenesis. Infect Immun 1999; 67:6385–6393.
Miao EA, Scherer CA, Tsolis RM, et al. Salmonella typhimurium leucine-rich repeat proteins are targeted to the SPI1 and SPI2 type III secretion systems. Mol Microbiol 1999; 34:850–864.
Bernal-Bayard J, Ramos-Morales F . Salmonella type III secretion effector SlrP is an E3 ubiquitin ligase for mammalian thioredoxin. J Biol Chem 2009; 284:27587–27595.
Bernal-Bayard J, Cardenal-Munoz E, Ramos-Morales F . The Salmonella type III secretion effector, Salmonella leucine-rich repeat protein (SlrP), targets the human chaperone ERdj3. J Biol Chem 2010; 285:16360–16368.
Haraga A, Miller SI . A Salmonella type III secretion effector interacts with the mammalian serine/threonine protein kinase PKN1. Cell Microbiol 2006; 8:837–846.
Auweter SD, Bhavsar AP, de Hoog CL, et al. Quantitative mass spectrometry catalogues Salmonella pathogenicity island-2 effectors and identifies their cognate host binding partners. J Biol Chem 2011; 286:24023–24035.
Bhavsar AP, Brown NF, Stoepel J, et al. The Salmonella type III effector SspH2 specifically exploits the NLR co-chaperone activity of SGT1 to subvert immunity. PLoS Pathog 2013; 9:e1003518.
Wiesner S, Ogunjimi AA, Wang HR, et al. Autoinhibition of the HECT-type ubiquitin ligase Smurf2 through its C2 domain. Cell 2007; 130:651–662.
Leung KY, Reisner BS, Straley SC . YopM inhibits platelet aggregation and is necessary for virulence of Yersinia pestis in mice. Infect Immun 1990; 58:3262–3271.
Evdokimov AG, Anderson DE, Routzahn KM, Waugh DS . Unusual molecular architecture of the Yersinia pestis cytotoxin YopM: a leucine-rich repeat protein with the shortest repeating unit. J Mol Biol 2001; 312:807–821.
Soundararajan V, Patel N, Subramanian V, Sasisekharan V, Sasisekharan R . The many faces of the YopM effector from plague causative bacterium Yersinia pestis and its implications for host immune modulation. Innate Immun 2011; 17:548–557.
McDonald C, Vacratsis PO, Bliska JB, Dixon JE . The Yersinia virulence factor YopM forms a novel protein complex with two cellular kinases. J Biol Chem 2003; 278:18514–18523.
Höfling S, Grabowski B, Norkowski S, Schmidt MA, Rüter C . Current activities of the Yersinia effector protein YopM. Int J Med Microbiol 2015; 305:424–432.
LaRock CN, Cookson BT . The Yersinia virulence effector YopM binds caspase-1 to arrest inflammasome assembly and processing. Cell Host Microbe 2012; 12:799–805.
Cohen P, Tcherpakov M . Will the ubiquitin system furnish as many drug targets as protein kinases? Cell 2010; 143:686–693.
Murray MF, Jurewicz AJ, Martin JD, et al. A high-throughput screen measuring ubiquitination of p53 by human mdm2. J Biomol Screen 2007; 12:1050–1058.
Hong CA, Swearingen E, Mallari R, et al. Development of a high throughput time-resolved fluorescence resonance energy transfer assay for TRAF6 ubiquitin polymerization. Assay Drug Dev Technol 2003; 1:175–180.
Orlicky S, Tang X, Neduva V, et al. An allosteric inhibitor of substrate recognition by the SCFCdc4 ubiquitin ligase. Nat Biotechnol 2010; 28:733–737.
Varfolomeev E, Blankenship JW, Wayson SM, et al. IAP antagonists induce autoubiquitination of c-IAPs, NF-κB activation, and TNFα-dependent apoptosis. Cell 2007; 131:669–681.
Vassilev LT, Vu BT, Graves B, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 2004; 303:844–848.
Rossi M, Rotblat B, Ansell K, et al. High throughput screening for inhibitors of the HECT ubiquitin E3 ligase ITCH identifies antidepressant drugs as regulators of autophagy. Cell Death Dis 2014; 5:e1203.
Maculins T, Carter N, Dorval T, et al. A generic platform for cellular screening against ubiquitin ligases. Sci Rep 2016; 6:18940.
Uetz P, Dong YA, Zeretzke C, et al. Herpesviral protein networks and their interaction with the human proteome. Science 2006; 311:239–242.
Mukhtar MS, Carvunis AR, Dreze M, et al. Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science 2011; 333:596–601.
Hunter T . The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol Cell 2007; 28:730–738.
Wijdeven RH, Neefjes J, Ovaa H . How chemistry supports cell biology: the chemical toolbox at your service. Trends Cell Biol 2014; 24:751–760.
Ovaa H . Active-site directed probes to report enzymatic action in the ubiquitin proteasome system. Nat Rev Cancer 2007; 7:613–620.
An H, Statsyuk AV . Development of activity-based probes for ubiquitin and ubiquitin-like protein signaling pathways. J Am Chem Soc 2013; 135:16948–16962.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 Unported License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Maculins, T., Fiskin, E., Bhogaraju, S. et al. Bacteria-host relationship: ubiquitin ligases as weapons of invasion. Cell Res 26, 499–510 (2016). https://doi.org/10.1038/cr.2016.30
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2016.30
Keywords
This article is cited by
-
An expanded lexicon for the ubiquitin code
Nature Reviews Molecular Cell Biology (2023)
-
IL-1β turnover by the UBE2L3 ubiquitin conjugating enzyme and HECT E3 ligases limits inflammation
Nature Communications (2023)
-
Chromosome-encoded IpaH ubiquitin ligases indicate non-human enteroinvasive Escherichia
Scientific Reports (2022)
-
Gasdermin B in the host–pathogen tug-of-war
Cell Research (2021)
-
Structural basis for protein glutamylation by the Legionella pseudokinase SidJ
Nature Communications (2021)