Abstract
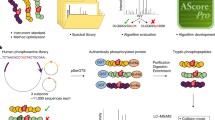
Reversible protein phosphorylation has been known for some time to control a wide range of biological functions and activities1,2,3. Thus determination of the site(s) of protein phosphorylation has been an essential step in the analysis of the control of many biological systems. However, direct determination of individual phosphorylation sites occurring on phosphoproteins in vivo has been difficult to date, typically requiring the purification to homogeneity of the phosphoprotein of interest before analysis4,5,6. Thus, there has been a substantial need for a more rapid and general method for the analysis of protein phosphorylation in complex protein mixtures. Here we describe such an approach to protein phosphorylation analysis. It consists of three steps: (1) selective phosphopeptide isolation from a peptide mixture via a sequence of chemical reactions, (2) phosphopeptide analysis by automated liquid chromatography–tandem mass spectrometry (LC-MS/MS), and (3) identification of the phosphoprotein and the phosphorylated residue(s) by correlation of tandem mass spectrometric data with sequence databases. By utilizing various phosphoprotein standards and a whole yeast cell lysate, we demonstrate that the method is equally applicable to serine-, threonine- and tyrosine-phosphorylated proteins, and is capable of selectively isolating and identifying phosphopeptides present in a highly complex peptide mixture.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Graves, J.D. & Krebs, E.D. Protein phosphorylation and signal transduction. Pharmacol. Ther. 82, 111–121 (1999).
Koch, C.A., Anderson, D., Moran, M. F., Ellis, C. & Pawson, T. SH2 and SH3 domains: elements that control interactions of cytoplasmic signaling proteins. Science 252, 668–674 (1991).
Hunter, T. 1001 protein kinases redux—towards 2000. Semin. Cell Biol. 5, 367–376 (1994).
Verma, R. et al. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science 278, 455–460 (1997).
Watts, J.D. et al. Identification by electrospray ionization mass spectrometry of the sites of tyrosine phosphorylation induced in activated Jurkat T cells on the protein tyrosine kinase ZAP-70. J. Biol. Chem. 269, 29520–29529 (1994).
Gingras, A.C. et al. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 13, 1422–1437 (1999).
Bodanszky, A.B.M. (ed.) The practice of peptide synthesis, Vol. 21. (Springer-Verlag, New York; 1984).
Hoare, D.G. & Koshland, D.E. Jr. A method for the quantitative modification and estimation of carboxylic acid groups in proteins. J. Biol. Chem. 242, 2447–2453 (1967).
Chu, B., Wahl, G.M. & Orgel, L.E. Derivatization of unprotected polynucleotides. Nucleic Acids Res. 11, 6513–6529 (1983).
Papayannopoulos, I.A. The interpretation of collision-induced dissociation tandem mass spectra of peptides. Mass Spectrom. Rev. 14, 49–73 (1995).
Jonscher, K.R. & Yates, J.R. Matrix-assisted laser desorption ionization/quadrupole ion trap mass spectrometry of peptides. Application to the localization of phosphorylation sites on the P protein from Sendai virus. J. Biol. Chem. 272, 1735–1741 (1997).
Qin, J. & Chait, B.T. Identification and characterization of posttranslational modifications of proteins by MALDI ion trap mass spectrometry. Anal. Chem. 69, 4002–4009 (1997).
Aebersold, R., Watts, J.D., Morrison, H.D. & Bures, E.J. Determination of the site of tyrosine phosphorylation at the low picomole level by automated solid-phase sequence analysis. Anal. Biochem. 199, 51–60. (1991).
Gygi, S.P. et al. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 17, 994–999 (1999).
Gygi, S.P., Rochon, Y., Franza, B.R. & Aebersold, R. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19, 1720–1730 (1999).
Futcher, B., Latter, G.I., Monardo, P., McLaughlin, C.S., Garrels, J.I. A sampling of the yeast proteome. Mol. Cell. Biol. 19, 7357–7368 (1999).
Moore, D.D. et al. (eds). Current protocols in molecular biology. (Wiley, New York; 1987).
Carraway, K.L. & Koshland, D.E. Jr. Reaction of tyrosine residues in proteins with carbodiimide reagents. Biochim. Biophys. Acta 160, 272–274 (1968).
Eng, J., McCormack, A.L. & Yates, J.R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 (1994).
Acknowledgements
The National Science Foundation Science and Technology Center for Molecular Biotechnology, the National Institutes of Health (RO1 A141109 and 1R33 CA84698), the NIH Research Resource Center (RR11823), and the Merck Genome Research Institute provided support for this work. We thank Drs. Beate Rist, Steven P. Gygi, and David R. Goodlett for helpful discussions.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhou, H., Watts, J. & Aebersold, R. A systematic approach to the analysis of protein phosphorylation. Nat Biotechnol 19, 375–378 (2001). https://doi.org/10.1038/86777
Issue Date:
DOI: https://doi.org/10.1038/86777