Abstract
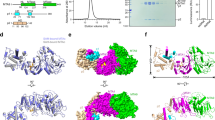
The 2.0 Å crystal structure of the N6-adenine DNA methyltransferase M•TaqI in complex with specific DNA and a nonreactive cofactor analog reveals a previously unrecognized stabilization of the extrahelical target base. To catalyze the transfer of the methyl group from the cofactor S-adenosyl-l-methionine to the 6-amino group of adenine within the double-stranded DNA sequence 5′-TCGA-3′, the target nucleoside is rotated out of the DNA helix. Stabilization of the extrahelical conformation is achieved by DNA compression perpendicular to the DNA helix axis at the target base pair position and relocation of the partner base thymine in an interstrand π-stacked position, where it would sterically overlap with an innerhelical target adenine. The extrahelical target adenine is specifically recognized in the active site, and the 6-amino group of adenine donates two hydrogen bonds to Asn 105 and Pro 106, which both belong to the conserved catalytic motif IV of N6-adenine DNA methyltransferases. These hydrogen bonds appear to increase the partial negative charge of the N6 atom of adenine and activate it for direct nucleophilic attack on the methyl group of the cofactor.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Cheng, X. Annu. Rev. Biophys. Biomol. Struct. 24, 293–318 (1995).
Jost, J.P. & Saluz, H.P. DNA methylation: molecular biology and biological significance (Birkhäuser Verlag, Basel; 1993).
Wu, J.C. & Santi, D.V. J. Biol. Chem. 262, 4778–4786 (1987).
Klimasauskas, S., Kumar, S., Roberts, R.J. & Cheng, X. Cell 76, 357–369 (1994).
Reinisch, K.M., Chen, L., Verdine, G.L. & Lipscomb, W.N. Cell 82, 143–153 (1995).
Malone, T., Blumenthal, R.M. & Cheng, X. J. Mol. Biol. 253, 618–632 (1995).
Pósfai, J., Bhagwat, A.S., Pósfai, G. & Roberts, R.J. Nucleic Acids Res. 17, 2421–2435 (1989).
Pogolotti, A.L., Ono, A., Subramaniam, R. & Santi, D.V. J. Biol. Chem. 263, 7461–7464 (1988).
Ho, D.K., Wu, J.C., Santi, D.V. & Floss, H.G. Arch. Biochem. Biophys. 284, 264–269 (1991).
Holz, B. et al. J. Biol. Chem. 274, 15066–15072 (1999).
Holz, B. & Weinhold, E. In Bioorganic chemistry: highlights and new aspects (eds, Diederichsen, U., Lindhorst, T.K., Westermann B. & Wessjohann, L.) 337–345 (Wiley-VCH, Weinheim; 1999).
Labahn, J. et al. Proc. Natl. Acad. Sci. USA 91, 10957–10961 (1994).
Tran, P.H., Korszun, Z.R., Cerritelli, S., Springhorn, S.S. & Lacks, S.A. Structure 6, 1563–1575 (1998).
Gong, W., O'Gara, M., Blumenthal, R.M. & Cheng, X. Nucleic Acids Res. 25, 2702–2715 (1997).
Schluckebier, G., Kozak, M., Bleimling, N., Weinhold, E. & Saenger, W. J. Mol. Biol. 265, 56–67 (1997).
Vassylyev, D.G. et al. Cell 83, 773–782 (1995).
Slupphaug, G. et al. Nature 384, 87–92 (1996).
Barrett, T.E. et al. Cell 92, 117–129 (1998).
Lau, A.Y., Schärer, O.D., Samson, L., Verdine, G.L. & Ellenberger, T. Cell 95, 249–258 (1998).
Hosfield, D.J., Guan, Y., Haas, B.J., Cunningham, R.P. & Tainer, J.A. Cell 98, 397–408 (1999).
Bruner, S.D., Norman, D.P.G. & Verdine, G.L. Nature 403, 859–866 (2000).
Cheng, X., Kumar, S., Posfai, J., Pflugrath, J.W. & Roberts, R.J. Cell 74, 299–307 (1993).
Schluckebier, G., O'Gara, M., Saenger, W. & Cheng, X. J. Mol. Biol. 247, 16–20 (1995).
Kettani, A., Guéron, M. & Leroy, J.-L. J. Am. Chem. Soc. 119, 1108–1115 (1997).
Schluckebier, G., Labahn, J., Granzin, J. & Saenger, W. Biol. Chem. 379, 389–400 (1998).
Arnett, E.M. Progr. Phys. Org. Chem. 1, 223–403 (1963).
Holz, B., Klimasauskas, S., Serva, S. & Weinhold, E. Nucleic Acids Res. 26, 1076–1083 (1998).
Kabsch, W. J. Appl. Cryst. 26, 795–800 (1993).
Navaza, J. Acta Crystallogr. A 50, 157–163 (1994).
Jones, T.A., Zou, J.-Y., Cowan, S.W. & Kjeldgaard, M. Acta Crystallogr. A 47, 110–119 (1991).
Brünger, A.T. et al. Acta Crystallogr. D 54, 905–921 (1998).
Acknowledgements
We would like to thank N. Bleimling for producing M•TaqI, S. Milardovic′ for help during initial crystallization experiments and M. Weyand for help with the refinement and graphical presentation. This work was supported in part by a grant from the Deutsche Forschungsgemeinschaft.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Goedecke, K., Pignot, M., Goody, R. et al. Structure of the N6-adenine DNA methyltransferase M•TaqI in complex with DNA and a cofactor analog. Nat Struct Mol Biol 8, 121–125 (2001). https://doi.org/10.1038/84104
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/84104