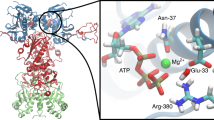
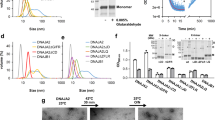
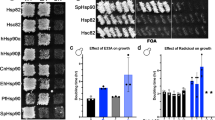
Abstract
The 70 kDa heat shock proteins (the Hsp70 family) assist refolding of their substrates through ATP-controlled binding. We have analyzed mutants of DnaK, an Hsp70 homolog, altered in key residues of its substrate binding domain. Substrate binding occurs by a dynamic mechanism involving: a hydrophobic pocket for a single residue that is crucial for affinity, a two-layered closing device involving independent action of an α-helical lid and an arch, and a superimposed allosteric mechanism of ATP-controlled opening of the substrate binding cavity that operates largely through a β-structured subdomain. Correlative evidence from mutational analysis suggests that the ADP and ATP states of DnaK differ in the frequency of the conformational changes in the α-helical lid and β-domain that cause opening of the substrate binding cavity. The affinity for substrates, as defined by this mechanism, determines the efficiency of DnaJ-mediated and ATP hydrolysis mediated locking-in of substrates and chaperone activity of DnaK.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Buchberger, A. et al. Nucleotide-induced conformational changes in the ATPase and substrate binding domains of the DnaK chaperone provide evidence for interdomain communication. J. Biol. Chem. 270, 16903–16910 (1995).
Schmid, D., Baici, A., Gehring, H. & Christen, P. Kinetics of molecular chaperone action. Science 263, 971–973 (1994).
Bukau, B. & Horwich, A.L. The Hsp70 and Hsp60 chaperone machines. Cell 92, 351–366 (1998).
Karzai, A.W. & McMacken, R. A bipartite signaling mechanism involved in DnaJ-mediated activation of the Escherichia coli DnaK protein. J. Biol. Chem. 271, 11236–11246 (1996).
Laufen, T. et al. Mechanism of regulation of Hsp70 chaperones by DnaJ co-chaperones. Proc. Natl. Acad. Sci. USA 96, 5452–5457 (1999).
Misselwitz, B., Staeck, O. & Rapoport, T.A. J proteins catalytically activate Hsp70 molecules to trap a wide range of peptide sequences. Mol. Cell 2, 593–603 (1998).
Zhu, X. et al. Structural analysis of substrate binding by the molecular chaperone DnaK. Science 272, 1606–1614 (1996).
Morshauser, R.C. et al. High-resolution solution structure of the 18 kDa substrate-binding domain of the mammalian chaperone protein Hsc70. J. Mol. Biol. 289, 1387–1403 (1999).
Wang, H. et al. NMR solution structure of the 21 kDa chaperone protein DnaK substrate binding domain: a preview of chaperone–protein interaction. Biochemistry 37, 7929–7940 (1998).
Pellecchia, M. et al. Structural insights into substrate binding by the molecular chaperone DnaK. Nature Struct. Biol. 7, 298–303 (2000).
Matlack, K.E.S., Misselwitz, B., Plath, K. & Rapoport, T.A. BiP acts as a molecular ratchet during posttranslational transport of prepro-α factor across the ER membrane. Cell 97, 553–564 (1999).
Vriend, G. A molecular modeling and drug design program. J. Mol. Graph. 8, 52–56 (1990).
McCarty, J.S. et al. Regulatory region C of the E. coli heat shock transcription factor, α32, constitutes a DnaK binding site and is conserved among eubacteria. J. Mol. Biol. 256, 829–837 (1996).
Flynn, G.C., Chappell, T.G. & Rothman, J.E. Peptide binding and release by proteins implicated as catalysts of protein assembly. Science 245, 385–390 (1989).
Takeda, S. & McKay, D.B. Kinetics of peptide binding to the bovine 70 kDa heat shock cognate protein, a molecular chaperone. Biochemistry 35, 4636–4644 (1996).
Greene, L.E., Zinner, R., Naficy, S. & Eisenberg, E. Effect of nucleotide on the binding of peptides to 70-kDa heat shock protein. J. Biol. Chem. 270, 2967–2973 (1995).
Pierpaoli, E.V., Gisler, S.M. & Christen, P. Sequence-specific rates of interaction of target peptides with the molecular chaperones DnaK and DnaJ. Biochemistry 37, 16741–16748 (1998).
Rüdiger, S., Germcroth, L., Schneider-Mergener, J. & Bukau, B. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 16, 1501–1507 (1997).
Bukau, B. & Walker, G.C. ΔdnaK mutants of Escherichia coli have defects in chromosomal segregation and plasmid maintenance at normal growth temperatures. J. Bacteriol. 171, 6030–6038 (1989).
Spence, J., Cegielska, A. & Georgopoulos, C. Role of Escherichia coli heat shock proteins DnaK and HtpG (C62.5) in response to nutritional deprivation. J. Bacteriol. 172, 7157–7166 (1990).
Ungewickell, E., Ungewickell, H. & Holstein, S.E.H. Functional interaction of the auxilin J domain with the nucleotide- and substrate-binding modules of Hsc70. J. Biol. Chem. 272, 19594–19600 (1997).
Gamer, J. et al. A cycle of binding and release of the DnaK, DnaJ and GrpE chaperones regulates activity of the E. coli heat shock transcription factor σ32. EMBO J. 15, 607–617 (1996).
Schröder, H., Langer, T., Hartl, F.-U. & Bukau, B. DnaK, DnaJ, GrpE form a cellular chaperone machinery capable of repairing heat-induced protein damage. EMBO J. 12, 4137–4144 (1993).
Rüdiger, S. Buchberger, A. & Bukau, B. Interaction of Hsp70 chaperones with substrates. Nature Struct. Biol. 4, 342–349 (1997).
Stenberg, G. & Fersht, A.R. Folding of barnase in the presence of the molecular chaperone SecB. J. Mol. Biol. 274, 268–275 (1997).
Fekkes, P., den Blaauwen, T. & Driessen, A.J.M. Diffusion-limited interaction between unfolded polypeptides and the Escherichia coli chaperone SecB. Biochemistry 34, 10078–10085 (1995).
Kunkel, T.A., Bebenek, K. & McClary, J. Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 204, 125–139 (1991).
Buchberger, A., Schröder, H., Büttner, M., Valencia, A. & Bukau, B. A conserved loop in the ATPase domain of the DnaK chaperone is essential for stable binding of GrpE. Nature Struct. Biol. 1, 95–101 (1994).
Mayer, M.P., Laufen, T., Paal, K., McCarty, J.S. & Bukau, B. Investigation of the interaction between DnaK and DnaJ by surface plasmon resonance. J. Mol. Biol. 289, 1131–1144 (1999).
Theyssen, H., Schuster, H.-P., Bukau, B. & Reinstein, J. The second step of ATP binding to DnaK induces peptide release. J. Mol. Biol. 263, 657–670 (1996).
McCarty, J.S., Buchberger, A., Reinstein, J. & Bukau, B. The role of ATP in the functional cycle of the DnaK chaperone system. J. Mol. Biol. 249, 126–137 (1995).
Szabo, A. et al. The ATP hydrolysis-dependent reaction cycle of the Escherichia coli Hsp70 system-DnaK, DnaJ and GrpE. Proc. Natl. Acad. Sci. USA 91, 10345–10349 (1994).
Kraulis, J. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950 (1991).
Acknowledgements
We thank B. Krauβ for technical assistance and W. Haehnel and R. Loyal for peptide synthesis. This work was supported by grants from the DFG and the Fonds der Chemie to B.B.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mayer, M., Schröder, H., Rüdiger, S. et al. Multistep mechanism of substrate binding determines chaperone activity of Hsp70. Nat Struct Mol Biol 7, 586–593 (2000). https://doi.org/10.1038/76819
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/76819