Abstract
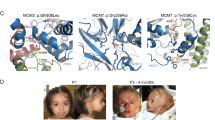
The recessive autosomal disorder known as ICF syndrome1,2,3 (for immunodeficiency, centromere instability and facial anomalies; Mendelian Inheritance in Man number 242860) is characterized by variable reductions in serum immunoglobulin levels which cause most ICF patients to succumb to infectious diseases before adulthood. Mild facial anomalies include hypertelorism, low-set ears, epicanthal folds and macroglossia. The cytogenetic abnormalities in lymphocytes are exuberant: juxtacentromeric heterochromatin is greatly elongated and thread-like in metaphase chromosomes, which is associated with the formation of complex multiradiate chromosomes. The same juxtacentromeric regions are subject to persistent interphase self-associations and are extruded into nuclear blebs or micronuclei. Abnormalities are largely confined to tracts of classical satellites 2 and 3 at juxtacentromeric regions of chromosomes 1, 9 and 16. Classical satellite DNA is normally heavily methylated at cytosine residues, but in ICF syndrome it is almost completely unmethylated in all tissues4. ICF syndrome is the only genetic disorder known to involve constitutive abnormalities of genomic methylation patterns. Here we show that five unrelated ICF patients have mutations in both alleles of the gene that encodes DNA methyltransferase 3B (refs 5, 6). Cytosine methylation is essential for the organization and stabilization of a specific type of heterochromatin, and this methylation appears to be carried out by an enzyme specialized for the purpose.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Maraschio,P., Zuffardi,O., Dalla Fior,T. & Tieplo,L. Immunodeficiency, centromeric heterochromatin instability of chromosomes 1, 9, and 16, and facial anomalies; the ICF syndrome. J. Med. Genet. 25, 173–180 (1988).
Hulten,M. Selective somatic pairing and fragility at 1q12 in a boy with common variable immunodeficiency. Clin. Genet. 14, 294 (1978).
Tieplo,L. et al. Concurrent instability at specific sites of chromosomes 1, 9, and 16 resulting in multibranched chromosomes. Clin. Genet. 14, 313–314 (1978).
Jeanpierre,M. et al. An embryonic-like methylation pattern of classical satellite DNA is observed in ICF syndrome. Hum. Mol. Genet. 2, 731–735 (1993).
Okano,M., Xie,S. & Li,E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5)-methyltransferases. Nature Genet. 19, 219–220 (1998).
Robertson,K. D. et al. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 27, 2291–2298 (1999).
Sumner,A. T., Mitchell,A. R. & Ellis,P. M. A FISH study of chromosome fusion in the ICF syndrome: involvement of paracentric heterochromatin but not of the centromeres themselves. J. Med. Genet. 35, 833–835 (1998).
Maraschio,P., Cortinovis,M., Dainotti,E., Tupler,R. & Tiepolo,L. Interphase cytogenetics of the ICF syndrome. Ann. Hum. Genet. 56, 273–288 (1992).
Lubit,B. W., Pham,T. D., Miller,O. J. & Erlanger,B. F. Localization of 5-methylcytosine in human metaphase chromosomes by immunoelectron microscopy. Cell 9, 503–509 (1976).
Yoder,J. A., Walsh,C. P. & Bestor,T. H. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13, 335–340 (1997).
Miniou,P. et al. Abnormal methylation pattern in constitutive and facultative (X inactive chromosome) heterochromatin of ICF patients. Hum. Mol. Genet. 3, 2093–2102 (1994).
Bourc'his,D. et al. Abnormal methylation does not prevent X inactivation in ICF patients. Cytogenet. Cell Genet. 84, 245–252 (1999).
Wijmenga,C. et al. Localization of the ICF syndrome to chromosome 20 by homozygosity mapping. Am. J. Hum. Genet. 63, 803–809 (1998).
Schuffenhauer,S. et al. DNA, FISH and complementation studies in ICF syndrome: DNA hypomethylation of repetitive and single copy loci and evidence for a trans acting factor. Hum. Genet. 96, 562–571 (1995).
Schuler,G. D. Pieces of the puzzle: expressed sequence tags and the catalog of human genes. J. Mol. Med. 75, 694–698 (1997).
Holliday,R. & Pugh,J. DNA modification mechanisms and gene activity during development. Science 187, 226–232 (1975).
Posfai,J., Bhagwat,A. S., Posfai,G. & Roberts,R. J. Predictive motifs derived from cytosine methyltransferases. Nucleic Acids Res. 17, 2421–2435 (1989).
Lauster,R., Trautner,T. A. & Noyer-Weidner,M. Cytosine-specific type II DNA methyltransferases. A conserved enzyme core with variable target-recognition domains. J. Mol. Biol. 206, 305–312 (1989).
Klimasauskas,S., Kumar,S., Roberts,R. J. & Cheng,S. HhaI methyltransferase flips its target base out of the DNA helix. Cell 76, 357–369 (1994).
Gibbons,R. J., Picketts,D. J., Villard,L. & Higgs,D. R. Mutations in a putative global transcriptional regulator cause X-linked mental retardation with athalassemia (ATR-X syndrome). Cell 80, 837–845 (1995).
Stec,I. et al. WHSC1, a 90 kb SET domain-containing gene, expressed in early development and homologous to a Drosophila dysmorphy gene maps in the Wolf–Hirschorn syndrome critical region and is fused to IgH in t(4;14) multiple myeloma. Hum. Mol. Genet. 7, 1071–1082 (1998).
Jeanpierre,M. Human satellites 2 and 3. Ann. Genet. 37, 163–171 (1994).
Hsieh,C.-L. In vivo activity of murine de novo methyltransferases, Dnmt3a and Dnmt3b. Mol. Cell. Biol. (in the press).
Jeddeloh,J. A., Stokes,T. L. & Richards,E. Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nature Genet. 22, 94–97 (1999).
Brown,K. E. et al. Association of transcriptionally silent genes with Ikaros complexes at contromeric heterochromatin. Cell 91, 845–854 (1997).
Chen,R. Z., Petterson,U., Beard,C., Jackson-Grusby,L. & Jaenisch,R. DNA hypomethylation leads to elevated mutation rates. Nature 395, 89–93 (1998).
Bestor,T. H. The host defence function of genomic methylation patterns. Novartis Found. Symp. 214, 187–195 (1998).
Hsieh,C.-L. Dependence of transcriptional repression on CpG methylation density. Mol. Cell. Biol. 14, 5487–5494 (1994).
Reynaud,C. et al. Monitoring of urinary excretion of modified nucleosides in cancer patients using a set of six monoclonal antibodies. Cancer Lett. 61, 255–262 (1992).
Acknowledgements
We thank members of the European ICF Consortium for providing patient materials; B. F. Erlanger for discussions; E. Li for providing cDNA clone pMT3B for mouse Dnmt3B; B. Tycko for DNA samples; L. Nickelsen for technical assistance; K. Anderson for comments on the manuscript; and A. Niveleau for monoclonal antibody to m5C. Supported by grants from the NIH and the Leukemia Society of America (T.H.B.) and the Danish Research Councils and the Danish Cancer Society (N.T.).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Xu, GL., Bestor, T., Bourc'his, D. et al. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature 402, 187–191 (1999). https://doi.org/10.1038/46052
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/46052
This article is cited by
-
DNMT3B PWWP mutations cause hypermethylation of heterochromatin
EMBO Reports (2024)
-
Placental DNA methylation profile as predicting marker for autism spectrum disorder (ASD)
Molecular Medicine (2023)
-
SMCHD1 and LRIF1 converge at the FSHD-associated D4Z4 repeat and LRIF1 promoter yet display different modes of action
Communications Biology (2023)
-
Enhanced CD19 activity in B cells contributes to immunodeficiency in mice deficient in the ICF syndrome gene Zbtb24
Cellular & Molecular Immunology (2023)
-
Differential expression of m5C RNA methyltransferase genes NSUN6 and NSUN7 in Alzheimer’s disease and traumatic brain injury
Molecular Neurobiology (2023)